Figure 3.
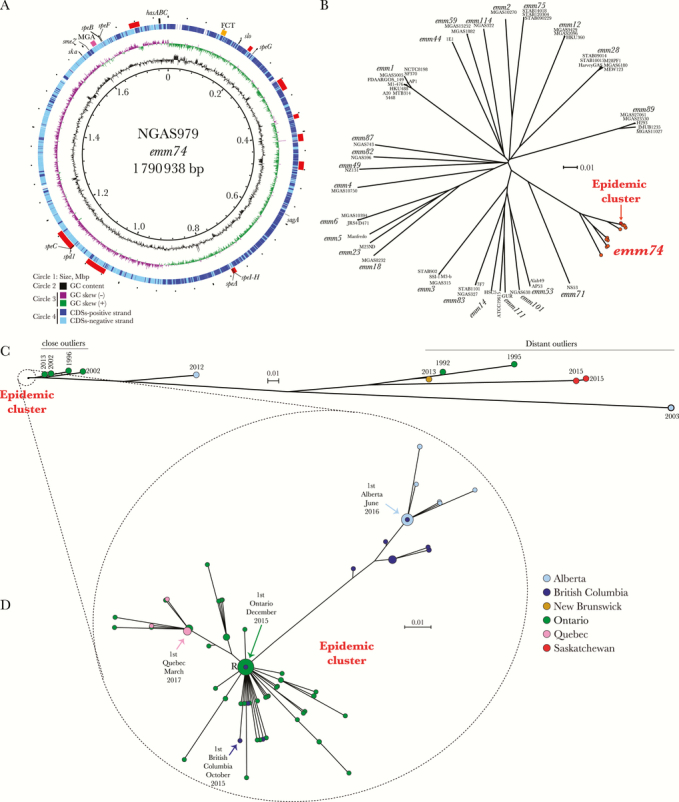
Genomic features and phylogenetic relationships of emm74 invasive group A Streptococcus isolates. A, Genome atlas of emm74 group A Streptococcus (GAS) reference strain NGAS979. Data from innermost to outermost circles in the atlas are described in the figure legend, with the exception of the outermost circle, which depicts genome landmarks such as mobile genetic element (prophages, prophage remnants, and integrative-conjugative elements, indicated by red boxes) and virulence genes, including those encoding superantigens. The hasABC locus is indicated by a black box, the FCT locus by a yellow box, and the mga regulon by a dark pink box. GC skew -or (G-C)/(G+C)- is averaged over a moving window of 10 000 bp. B, Inferred phylogenetic relationships between emm74 GAS strains and 55 strains of 25 other emm types, for which complete genome sequences are available in GenBank. A neighbor-joining phylogenetic tree was constructed using 76 311 nonredundant biallelic single nucleotide polymorphism (SNP) loci identified in the genomes of the strains relative to the core genome of the reference emm74 strain NGAS979. Strain names and emm types for non-emm74 strains are indicated at the tip of the branches. The reference emm74 strain and all 136 emm74 invasive and noninvasive isolates recovered since October 2015 in Canada clustered tightly in a discrete epidemic cluster (indicated in red). All other emm74 isolates from Canada are depicted in orange. C, Inferred phylogenetic relationship among 147 emm74 GAS strains from Canada. A neighbor-joining phylogenetic tree was constructed using 10 224 nonredundant biallelic SNP loci identified in the genomes of the emm74 isolates relative to the core genome of the reference emm74 strain NGAS979. The 136 emm74 GAS isolates recovered in Canada since October 2015 for which we generated genome data (the epidemic cluster) are monoclonal and genetically distinct from previously isolated emm74 strain SNPs. We arbitrarily divided these nonepidemic emm74 isolates into 3 groups: a first group of “close outliers,” comprising 4 isolates from Ontario in 1996, 2002, and 2013, a more distantly related single isolate from Alberta isolated in 2012, a group of 5 highly divergent “distant outlier” emm74 isolates recovered in Alberta, Saskatchewan, and Ontario from invasive infections, and 1 noninvasive isolate from New Brunswick recovered in 2013. Provinces are indicated by different colors, as per the caption in (D). D, Diversification of the emm74 GAS epidemic clone. The neighbor-joining phylogenetic tree was constructed using 71 nonredundant SNP loci identified among the 136 epidemic emm74 isolates relative to the genome of the reference strain NGAS979. The circles are colored to indicate province of isolation, as per the figure caption, and their sizes are proportional to the number of isolates with identical genotypes. The location in the phylogenetic tree of the first emm74 invasive isolate in each province is indicated by an arrow. Abbreviations: CDS, coding DNA sequence; GC, Guanine-cytosine.
