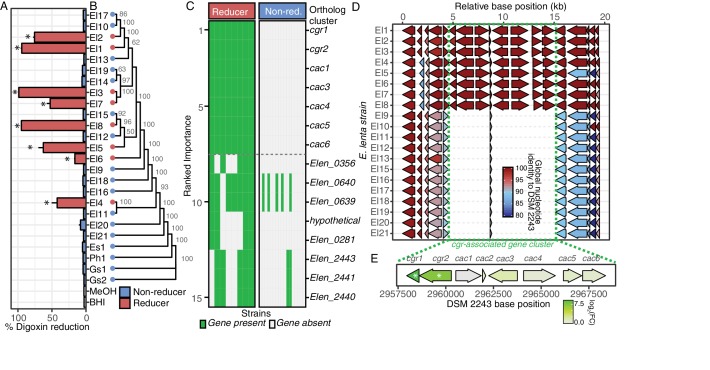
Figure 1. Comparative genomics expands the boundaries of the cgr operon.
(A) Survey of digoxin reduction in 21 strains of E. lenta (El#), 2 strains of Gordonibacter spp. (Gs#), E. sinensis (Es1), and Paraeggerthella hongkongesis (Ph1) (Figure 1—source data 1 and 2) revealed eight strains capable of reducing digoxin to dihydrodigoxin (*p<0.05, ANOVA with Dunnett’s test vs. vehicle controls). Data represents mean ± standard error of the mean (SEM) over three biological replicates. (B) Digoxin reduction did not correlate with phylogeny in E. lenta species (cladogram displayed with bootstrap values indicated at nodes; p=0.275, K = 0.049, Blomberg’s K). (C) Comparative genomics using a random forest classifier (see Materials and methods) revealed seven genes with perfect predictive accuracy for digoxin reduction. The orthologous cluster identified as hypothetical corresponds to an open reading frame present at position 299442..2995131 in the DSM 2243 reference genome. (D) Analysis of genomic context revealed a highly conserved 10.4 kb locus of 7 genes that flank a short, conserved hypothetical gene, herein termed the cgr-associated gene cluster (cac). (E) Analysis of gene expression in the cgr-associated gene cluster revealed only the cgr-locus was significantly upregulated by exposure to digoxin. * FDR < 0.1 (Figure 1—source data 3).