Figure 2. Cgr2 is sufficient for digoxin reduction and requires FAD and [4Fe-4S] cluster(s) for activity.
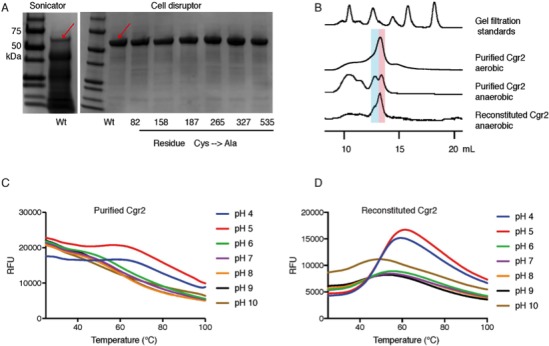
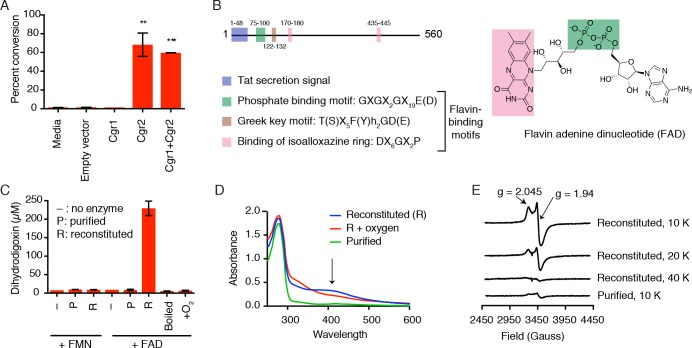
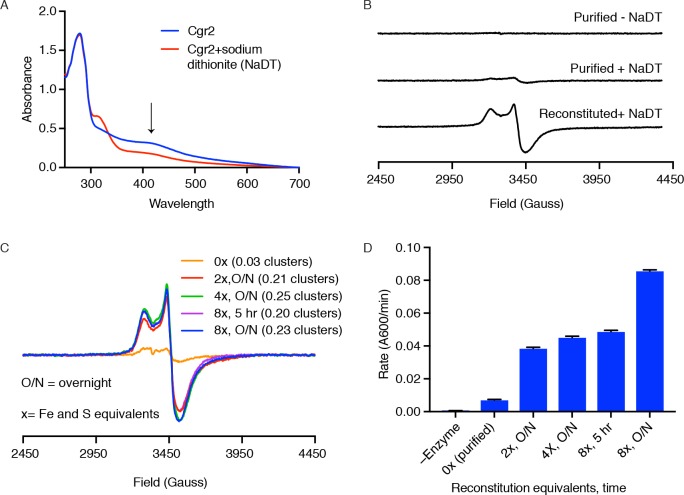
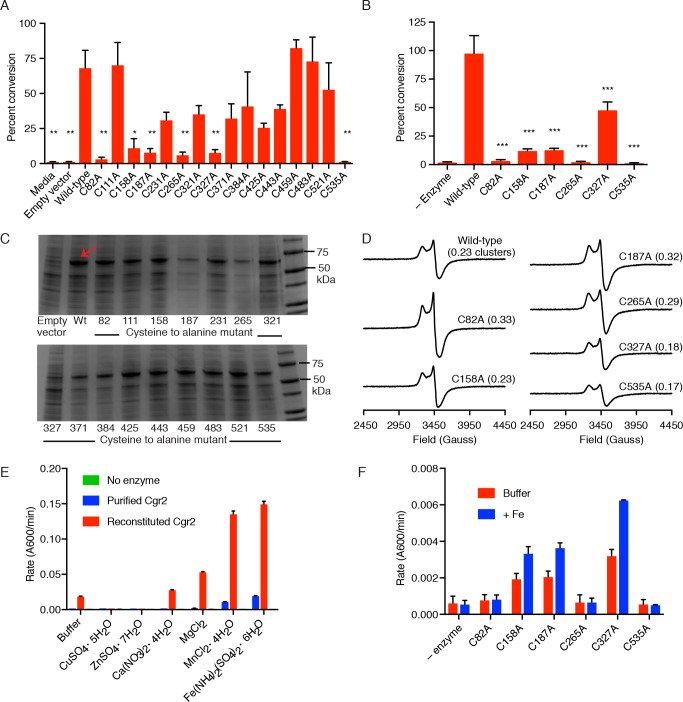
(A) Whole cell assays using R. erythropolis expressing Cgr1 and Cgr2 constructs demonstrated that Cgr2 is sufficient for reducing digoxin. Data represents the mean ± SEM (n = 3 biological replicates). Asterisks indicate statistical significance of each variant as compared to empty vector by Student’s t test (**p<0.01, ***p<0.001) (Figure 2—source data 1). (B) Annotation and amino acid numbering of Cgr2, including the predicted Tat secretion signal and three conserved flavin-binding motifs from the glutathione reductase family (X = any amino acid; h = hydrophobic residue). (C) In vitro activity of Cgr2 for digoxin reduction using reduced methyl viologen as an electron donor, analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). [Fe-S] cluster reconstitution, FAD, and anaerobic conditions are required for Cgr2 activity. Data represents the mean ± SEM (n = 3 independent experiments) (Figure 2—source data 2). FAD = flavin adenine dinucleotide; FMN = flavin mononucleotide. (D) Ultraviolet-visible (UV-Vis) absorption spectra of Cgr2 revealed an oxygen-sensitive peak centered around 400 nm that increased upon [Fe-S] cluster reconstitution, supporting the presence of [4Fe-4S] clusters in Cgr2. (E) Electron paramagnetic resonance (EPR) spectra of sodium dithionite-reduced Cgr2 reconstituted with iron ammonium sulfate hexahydrate ((NH4)2Fe(SO4)2·6H20) and sodium sulfide (Na2S·9H20). G-values and decreased EPR signal intensity at higher temperatures (10 – 40 K) indicated the presence of low potential [4Fe-4S]1+ clusters. Experimental conditions were microwave frequency 9.38 GHz, microwave power 0.2 mW, modulation amplitude 0.6 mT, and receiver gain 40 dB.
Figure 2—figure supplement 1. [Fe-S] cluster(s) affect Cgr2 stability and oligomerization.