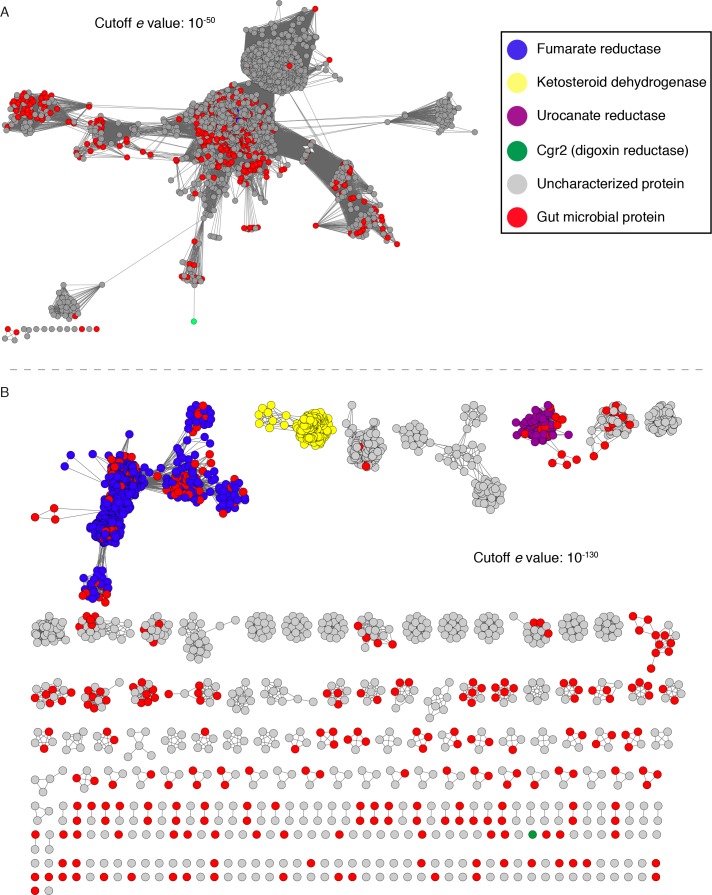
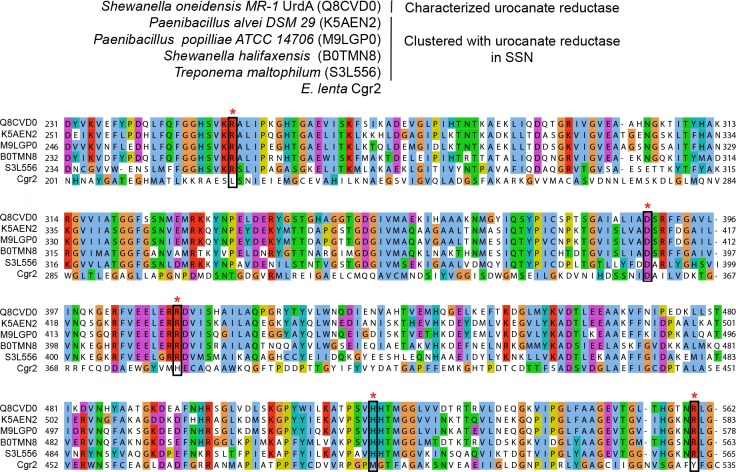
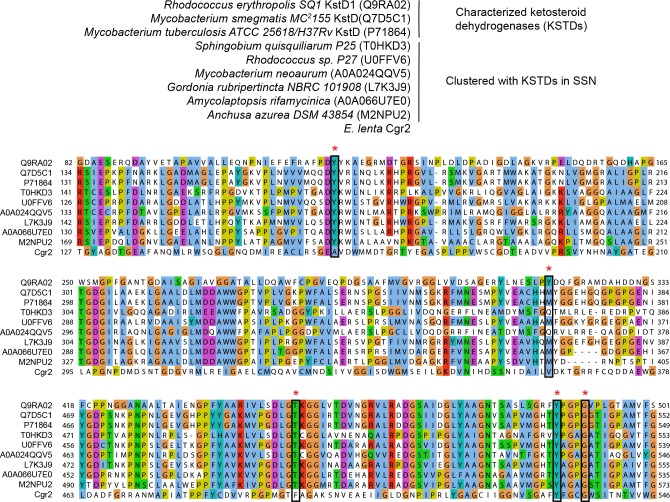
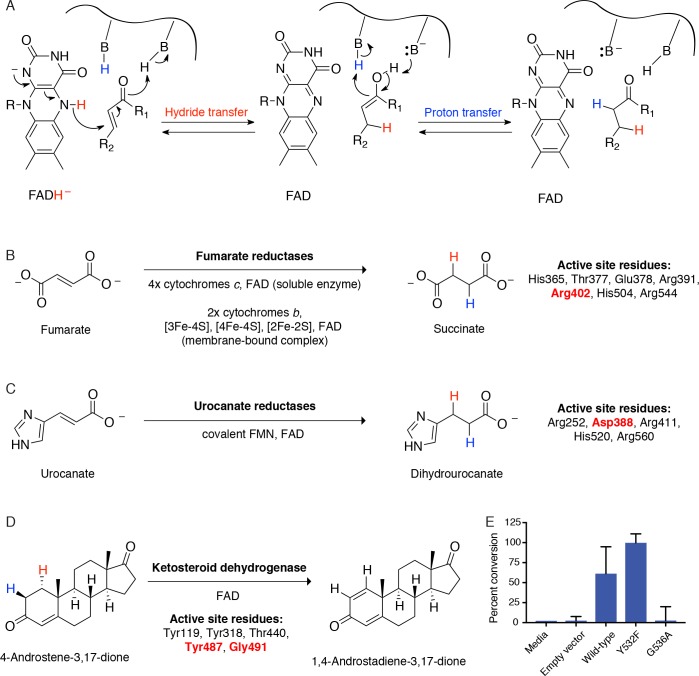
Figure 5. Sequence similarity network (SSN) analysis reveals that the gut bacterial enzyme Cgr2 is a highly distinct member of a large enzyme family that is widespread in gut microbes.
The SSN was constructed using the top 5000 most similar proteins to Cgr2 from the UniprotKB database. Nodes represent proteins with 100% sequence identity. (A) SSN displayed with an e-value threshold of 10−50. The seven previously characterized enzymes (PDB ID: 1D4D, 1E39; UniProtKB ID: Q07WU7, Q9Z4P0, 8CVD0, P71864, Q7D5C1) and Cgr2 are colored according to biochemical function. (B) SSN displayed with an e-value threshold of 10−130. All nodes that co-clustered with characterized enzymes are shown in the same color, denoting putative isofunctional activity. With the exception of Cgr2, if a node comes from a gut bacterium, it is colored red rather than the color of the corresponding cluster.