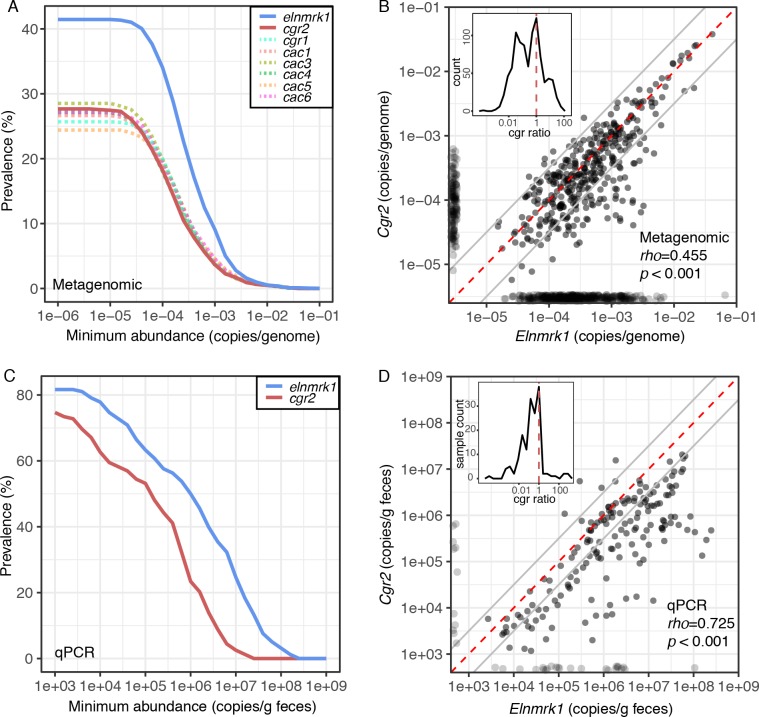
Figure 6. Cgr2 is widespread in the human gut microbiome.
(A) Analysis of the cgr-associated gene cluster and E. lenta (via elenmrk1) prevalence in the gut metagenomes of 1872 individuals (see Materials and methods) revealed that both E. lenta and cgr2 are highly prevalent (41.5% and 27.7% respectively) but frequently low in abundance. (B) Quantification of E. lenta and cgr2 abundances in individual gut metagenomes revealed a tight correlation between the two, providing evidence that cgr2 is restricted to E. lenta and that individuals may harbor sub-populations of both cgr2+ and cgr2- strains. Red line denotes the expected linear relationship and dashed lines represent a ± half log deviation. (Inset) Histogram of cgr-ratio (cgr/elnmrk1) demonstrates a significant skew away from communities that would have more cgr2 than expected by E. lenta abundance (p<0.001, D’Agostino skewness test). (C) Replication in an additional 158 individuals located in the USA (n = 85) and Germany (n = 73) via duplexed qPCR increased prevalence estimates to 74.7% and 81.6% at the extremes of detection limit (1e3 copies/g). qPCR samples were run in technical triplicate (Figure 6—source datas 1 and 2). (D) Similarly, qPCR-derived abundances of E. lenta and cgr2 were correlated, corroborating metagenome-based analysis. (Inset) Histogram of cgr-ratio demonstrating significant skew (p<0.001).