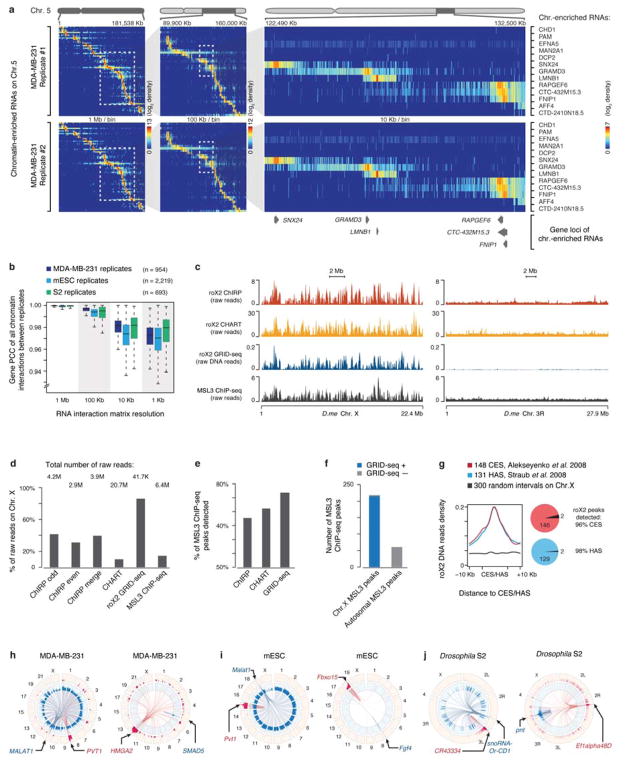
Extended Data Fig. 5. Characteristics of RNA-chromatin interactions.
a, RNA-chromatin interaction heatmaps on Chr. 5 constructed from two independent GRID-seq libraries on MDA-MB-231 cells. Boxed regions in each panel were enlarged with increasing resolution in the next panel on the right. A set of representative chromatin-enriched RNAs are labeled on the right and their encoding gene loci illustrated on the bottom. b, Pearson’s Correlation Coefficient of GRID-seq interaction density of each RNA at increasing resolution (decreasing bin size) across genomes between the replicates performed on MDA-MB-231 cells, mESC and Drosophila S2 cells. c, Whole chromosome view of raw roX2 GRID-seq DNA reads on Drosophila Chr. X and Chr. 3R in comparison with raw signals detected by ChIRP and CHART as well as ChIP-seq signals for the roX2 binding protein MSL3. d, Percentage of total raw roX2 GRID-seq DNA reads on Chr. X in comparison with the raw data of ChIRP, CHART, GRID-seq and MSL3 ChIP-seq. Total number of raw read for each dataset is noted on top of each bar. e, Comparison of roX2 peaks identified by ChIRP, CHART, and GRID-seq with MSL3 ChIP-seq peaks in Drosophila S2 cells, showing the highest overlap between GRID-seq and MSL3 ChIP-seq identified peaks. f, Comparison of GRID-seq identified roX2 peaks with MSL3 ChIP-seq peaks on Chr. X and autosomes, showing their overlap only on Chr. X. g, Meta-analysis of roX2 GRID-seq DNA reads relative to chromatin binding of CES and HAS. Pie charts on the right illustrate the overlaps between CES/HAS peaks and roX2 GRID-seq peaks. h,i,j, Circos plots depicting chromatin-interactions of two representing lncRNAs (left panels) and two protein-coding mRNAs (right panels) in each cell type. In MDA-MB-231 cells (h), the lncRNA MALAT1 broadly interacted with all chromosomal regions whereas PVT1 was predominantly involved in local and cis-chromosomal interactions, although trans-chromosomal interaction signals were also evident in various specific locations (h, left panel). Similarly, RNAs from the coding HMGA2 gene showed chromatin interactions in local, cis and trans modes whereas RNAs from the coding SMAD5 gene was largely confined in local interactions (h, right panel). Similar examples also illustrated distinct chromatin interaction patterns with both coding RNAs and lncRNAs in mESC (i) and Drosophila S2 cells (j).