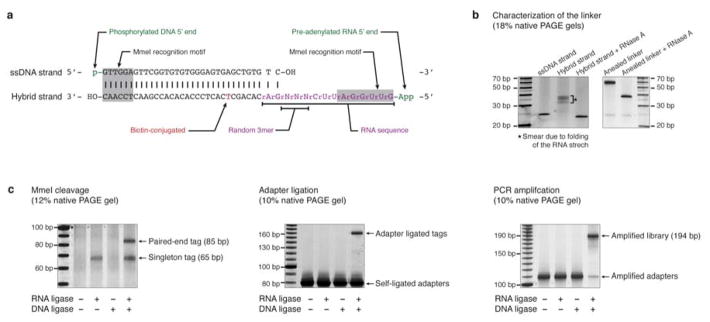
Extended Data Fig. 1. Characterization of the GRID-seq technology.
a, The design of a bivalent linker for GRID-seq. The top strand is a 5′ phosphorylated DNA sequence (black) and the bottom strand consists of both DNA and RNA bases (purple) with a biotinylated T residue (red) in the middle. Randomized bases (N) served as barcodes for filtering PCR duplicates during library amplification and both ends of the linker each carry an MmeI restriction site (grey-shaded). The linker is pre-adenylated for ligation to RNA in the absence of ATP, which prevents ligation of endogenous RNAs. b, Characterization of the linker before (left) and after (right) annealing, showing the sensitivity of the RNA-containing linker to RNase A. c, Controls by omitting RNA ligase, DNA ligase or both during GRID-seq library construction, which yielded expected ligated products of singleton tags or paired-end tags (left). After adapter ligation (middle) and PCR amplification (right), the expected products were only detected after library construction with both RNA and DNA ligases.