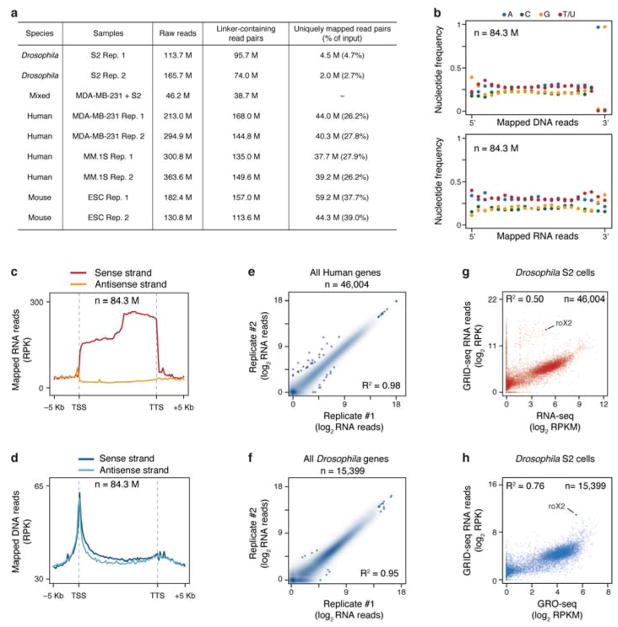
Extended Data Fig. 2. Characterization of GRID-seq libraries.
a, Summary of sequenced GRID-seq libraries constructed on two human cell lines (MDA-MB-231 and MM.1S), one mESC, and one Drosophila S2 cells. Shown are raw reads, linker-containing reads, and uniquely mapped reads from mated RNA/DNA pairs. b, Nucleotide frequency of DNA (up) and RNA (bottom) reads. Note specific dinucleotide as part of the AluI recognition site at the 3′ end of DNA reads, but the lack of nucleotide bias in any position of RNA reads. c,d, Strand orientation of mapped RNA (c) and DNA (d) reads. Note the same strand orientation of mapped RNA reads as their transcripts, but not DNA. e,f, Reproducibility of GRID-seq libraries constructed on human (e) and Drosophila (f) cells. g,h, Comparison of GRID-seq detected RNA reads with gene expression detected by RNA-seq of rRNA-depleted total RNA (g) or GRO-seq (h) in Drosophila S2 cells. The lncRNA roX2 is highlighted in both plots. RPK: GRID-seq reads per Kb. RPKM: reads per Kb per million mapped reads.