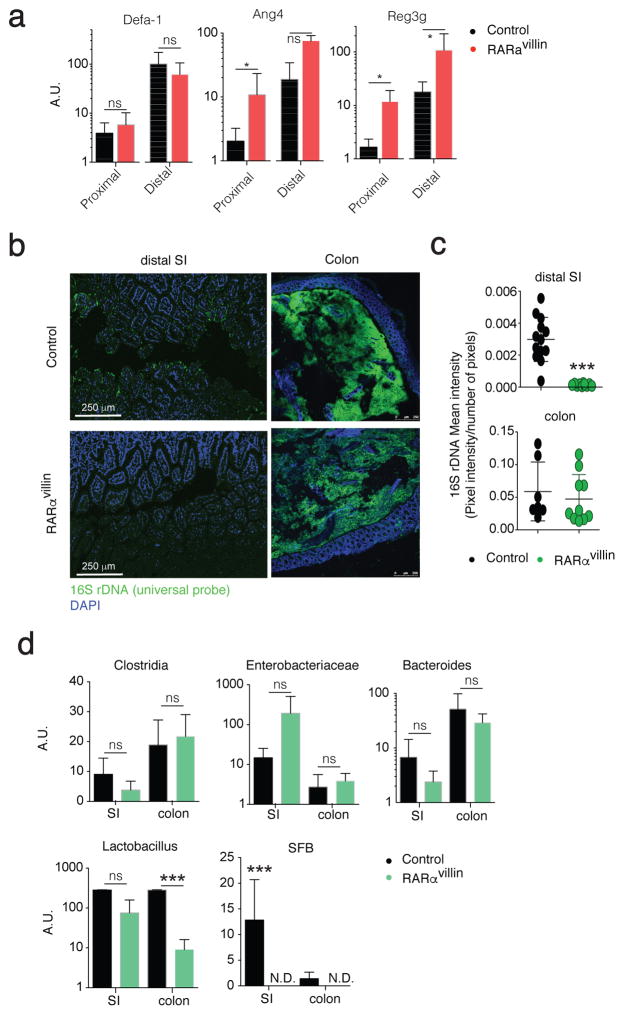
Figure 4.
Dysbiosis in RARαvillin mice. (a) qPCR analysis from FACS-sorted epithelial cells (CD45negEpCAM+) obtained from the proximal or distal small intestine of either control or RARαvillin mice. Data shows transcript levels as arbitrary units (A.U.) respect to hprt (n = 3 mice). (b) Fluorescence in situ hybridization of universal 16S ribosomal RNA in DAPI-stained ileal tissues from control and RARαvillin mice. Original magnification, 10X. One representative image of 3–6 images/mouse (n = 2 mice). (c) 16S rDNA mean intensities (sum of pixel intensities/number of pixels) are reported. Scale bar, 10 μm. (d) qPCR analysis shows arbitrary units (A.U.) of lactobacillus, enterobacterae (entero), bacteroides, clostridia and segmented filamentous bacteria (SFB) relative to universal 16S levels. Bacterial DNA was isolated from luminal stool obtained from the small intestine (SI) or colon (n = 3; 2 experiments). *P < 0.05; *** P < 0.005; ns, non-significant; N.D., non-detected; Student’s t-test. Results are shown as mean ± SEM in all panels. Scale bar; 250 μm (b)