FIGURE 1.
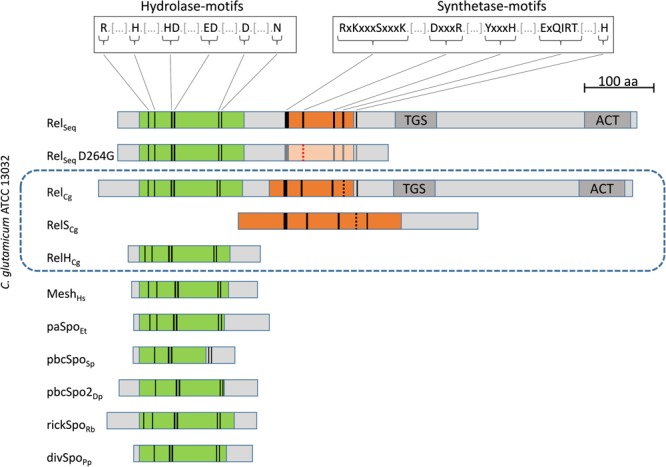
Comparative structural analysis of enzymes involved in (pp)pGpp metabolism. True-to-scale representation of domain architecture and conserved sequence motives of stringent response associated enzymes of C. glutamicum ATCC 13032 (enclosed by dashed blue lines) and further selected species. These include the native Rel enzyme of Streptococcus equisimilis RelSeq, as well as the shortened and only hydrolytically active variant RelSeq D264G (Hogg et al., 2004; Sun et al., 2010) and representatives of the six other SAH subgroups (Atkinson et al., 2011): Mesh (Homo sapiens), paSpo (Erwinia tasmaniensis), pbcSpo (S. pneumoniae), pbcSpo2 (Desulfotalea psychrophila), rickSpo (Rickettsia bellii), and divSpo (Photobacterium profundum). Hydrolase domain HD4 (pfam13328) and ppGpp synthetase catalytic domain YjbM (COG2357) are indicated in green and orange, respectively. Additional C-terminal elements of long RSH enzymes, which are likely to have regulatory functions and are classified as TGS (ThrRS, GTPase, and SpoT) and ACT (aspartate kinase, chorismate mutase and TyrA) domains (Wolf et al., 1999; Chipman and Shaanan, 2001), are also indicated. Matches with conserved synthetase and hydrolase motifs are represented by black lines (Bag et al., 2014; Steinchen and Bange, 2016). Minor deviations from the conserved sequence motifs are shown by dashed black lines. The amino acid exchange of the protein RelSeq D264G is represented by a red dashed line and the resulting non-functional YjbM domain is faded out.
