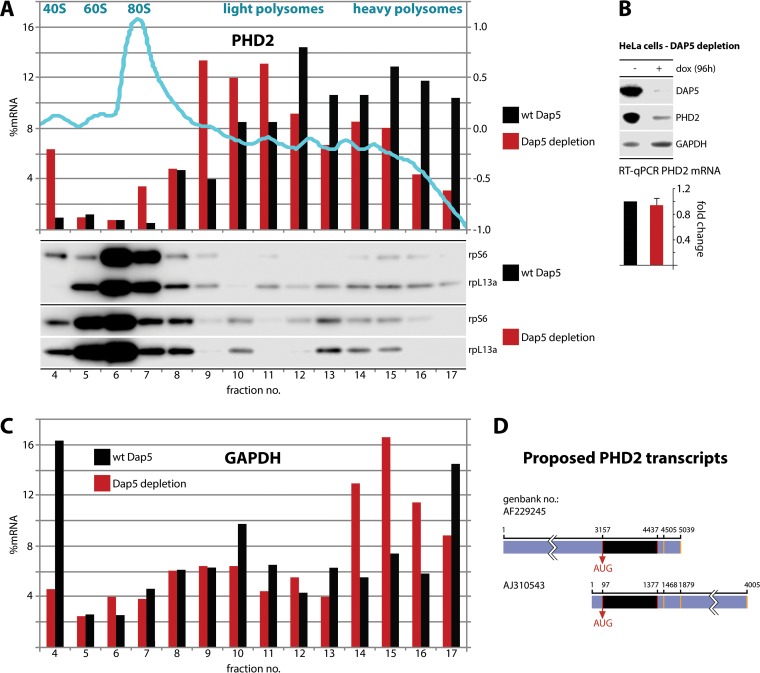
FIG 6.
DAP5 is involved in PHD2 translation. (A and C) HeLa cells with dox-inducible DAP5 depletion were dox treated (96 h), and lysates were subjected to polysome profiling. (Top panel) A representative absorbance (260:280) profile of the fractionated gradient is shown in light blue with the peaks identified. Total RNA was purified from the fractions, RT-qPCR analyses for PHD2 (A) and GAPDH (C) mRNA were performed, and the percentages of the PHD2/GAPDH mRNA signal were determined for each fraction. This experiment was repeated twice and yielded similar results in each series; representative profiles are shown. Proteins from each fraction were TCA precipitated and analyzed by immunoblotting. (B) HeLa cells with dox-inducible DAP5 depletion were treated with dox (96 h) or left untreated. Total polyadenylated RNA was isolated from the cells and analyzed by RT-qPCR. The average of two assays with the uninduced controls was normalized to 1, and the SEM values are shown. Representative immunoblots from lysates are shown. (D) Schematic view of the ambiguously annotated 3′ and 5′ UTRs of PHD2. Alternative transcriptional start sites and polyadenylation signals (orange bars) are indicated (see the text for references).