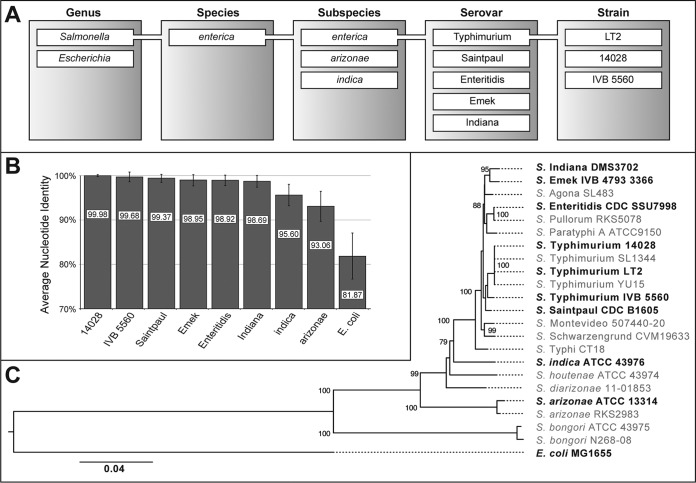
FIG 3 .
Phylogenetic relationships between selected host strains. (A) The host strain used in this study include representatives from different strains, serovars, subspecies, and genera to represent a broad spectrum of genetic backgrounds. (B) Average nucleotide identities (ANI) of all host strains versus S. Typhimurium LT2 range from 99.98% to 81.87%. For a complete comparison of ANI between all strains, see Table S4 in the supplemental material. (C) Maximum likelihood tree of the strains used in this study supplemented with other representatives of the Salmonella clade (rooted with E. coli). The tree was inferred from seven housekeeping genes (hisD, purE, sucA, thrA, aroC, dnaN, and hemD). Numbers on the branches represent the support values of 1,000 bootstrap replicates. Only values of >70 are presented. The bar indicates the number of substitutions per site.