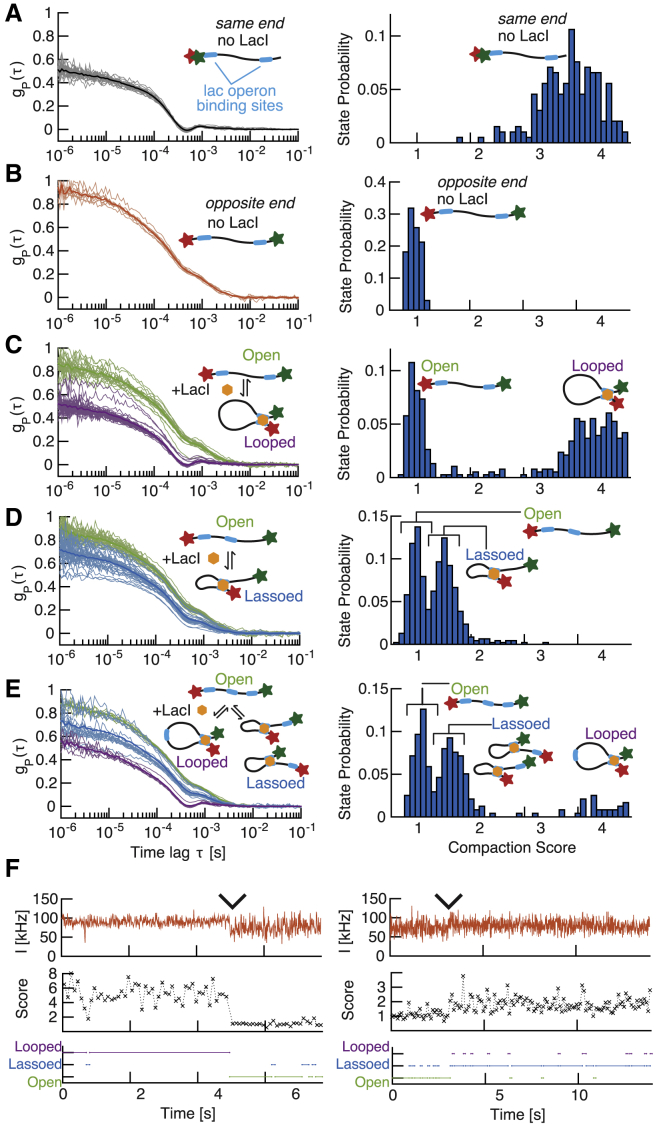
Figure 5.
Detection of protein-mediated conformational changes. (A and B) Left: single-molecule tFCS signals (thin lines) and population mean (thick lines) of the looping DNA construct bearing two lac operators are shown. The molecules were labeled with Atto647N and Cy3b either at the same end (A) or on opposite ends (B), and data are shown in the absence of LacI. Right: the distribution of compaction scores for the 100 ms subtraces of all the individual molecules is shown. (C–E) Left: tFCS signals of individual DNA molecules after addition of 64 nM LacI are shown. The DNA used were the looping construct in (C), the lasso construct in (D), and the construct with three LacO sites in (E). The conformational state inferred by clustering the tFCS signals are categorized as the following: open in green (no LacI bound or LacI bound at a single site), lassoed in blue (LacI-mediated short loop), or looped in purple (LacI-mediated long loop). Cluster averages (thick lines) and individual molecules (thin lines) are shown for each of these three groups. Right: the distribution of compaction scores for the 100 ms subtraces of all the individual molecules is shown. (F) A representative example of a conformational transition between a looped and an open state (left) or an open and a lassoed state (right), detected during the tracking of an individual molecule. Probe dye fluorescence trace (top), 100 ms binned compaction score (middle), and inferred conformational state (bottom) are shown. Wedges indicate time of transition between the two conformations. To see this figure in color, go online.