Abstract
Acetaminophen (APAP) overdose is the leading cause of acute liver failure (ALF) with limited treatment options. It is known that liver regeneration following APAP-induced ALF is a deciding factor in the final outcome. Previous studies from our laboratory using an incremental dose model involving a regenerating (300 mg/kg, APAP300) and a nonregenerating (600 mg/kg, APAP600) dose of APAP in mice have revealed several proregenerative pathways that regulate regeneration after APAP overdose. Here we report that DNA damage and repair mechanisms regulate initiation of liver regeneration following APAP overdose. Mice treated with nonregenerating APAP600 dose showed prolonged expression of pH2AX, a marker of the DNA double-strand break (DSB), compared with APAP300. In regenerating APAP300 dose-treated mice, H2AX was rapidly dephosphorylated at Tyr142, indicating timely DNA repair. Expression of several DNA repair proteins was substantially lower with APAP600. Poly(ADP) ribose polymerase (PARP) activation, involved in DNA repair, was significantly higher in the APAP300 group compared to the APAP600 group. Activation of p53, the major cell cycle checkpoint protein, was significantly higher with APAP600 as demonstrated by substantially higher expression of its target genes. Taken together, these data show that massive DNA DSB occurs in high-dose APAP toxicity, and lack of prompt DSB repair after APAP overdose leads to prolonged growth arrest and proliferative senescence, resulting in inhibited liver regeneration.
Key words: p53, Poly(ADP) ribose polymerase (PARP), pH2AX, Proliferation, Injury
INTRODUCTION
Acetaminophen (APAP) is a widely used analgesic and antipyretic drug present in several over-the-counter and prescription medications. It is safe at therapeutic doses of ≤4 g/day; however, overdose of APAP can cause acute liver injury (ALI), which can progress to acute liver failure (ALF)1. Overdose of APAP is the cause of almost 50% of ALF cases in the US, with close to 35% mortality2,3. Despite being the major cause of ALF in the Western world, therapeutic options for APAP-induced ALF are extremely limited. Several studies in patients and rodents have demonstrated that stimulation of liver regeneration improves survival and prognosis after APAP overdose4–9. Although these studies highlight enhancing liver regeneration in the APAP-induced ALF patients as a plausible therapeutic option, the clinical application is delayed because the mechanisms of liver regeneration that drive liver regeneration after APAP overdose are not entirely known. Especially, the role of DNA damage response (DDR) in the regulation of liver regeneration after APAP-induced ALI has not been investigated.
DDR involves proteins that sense DNA damage and trigger a repair response to protect the cell. Sensor proteins in DDR sense the damage and send the signal to mediator and effector proteins via activation of apical kinases. Mediator proteins recruit DNA repair effector proteins at the damaged DNA site, which then carry out the repair process10,11. One of the major effector proteins in DDR is p53, which activates cell cycle checkpoints and induces cell cycle arrest until damage is repaired. However, if damage is beyond repair it can activate the cell death pathway12. Previous studies have shown that APAP injury results in nuclear DNA fragmentation, preventing cell proliferation by cell cycle arrest5,13,14.
We investigated the role of DDR in liver regeneration after APAP toxicity using a recently developed incremental dose model in our laboratory that includes comparing signaling between a regenerating (300 mg/kg, APAP300) and a nonregenerating (600 mg/kg, APAP600) dose in mice5. Our studies indicate that APAP overdose results in dose-dependent DNA damage, but at higher doses the DNA repair mechanisms fail, resulting in initiation of cellular senescence and inhibition of liver regeneration. These studies have revealed a novel mechanism that connects cellular injury to initiation of liver regeneration after APAP overdose.
MATERIALS AND METHODS
Animals, Treatment, and Tissue Harvesting
All animal experiments were performed in compliance with protocols approved by the Institutional Animal Care and Use Committee at University of Kansas Medical Center. The details of the incremental dose model have been previously published5. Briefly, 2- to 3-month-old male C57BL/6 mice were fasted overnight and injected with either 300 or 600 mg/kg APAP intraperitoneally (IP; dissolved in warm saline). Mice (n = 5 to 7) were sacrificed at 0, 3, 6, 12, 24, 48, 72, and 96 h after APAP treatment, and blood and livers were collected. Parts of liver tissue were processed separately to obtain paraffin sections, frozen sections, RNA samples, and nuclear, cytoplasmic, and radioimmunoprecipitation assay (RIPA) total protein extracts as described previously15. Liver injury was assessed by serum alanine aminotransferase (ALT) activity. Liver regeneration was assessed using proliferating cell nuclear antigen (PCNA) analysis.
Antibodies
The following antibodies were used for analyses: KU70 (#4588), PARP (#9532), phosphorylated histone 2AX (pH2AX) Ser139 (#9718), p53 (#2524), and p-p53 S15 (#9284) (from Cell Signaling Technology, Danvers, MA, USA); DNAPkc (SC9051), KU80 (SC1485), XLF (SC166488), XRCC4 (SC8285), DNA Lig4 (SC28232), and BRCA1 (SC642) (from Santa Cruz Biotechnology, Santa Cruz, CA, USA); pH2Ax Tyr142 (#07-1590) (from EMD Millipore Corporation, Billerica, MA, USA); HNF4α (PP-H1415-00) (from Perseus Proteomics); 53BP1 (nb100-904) (from Novus Biologicals); and PAR (#1020) (from Tulips Biolabs). All Alexa Fluor secondary antibodies were purchased from Invitrogen, Thermo Fisher, and HRP-conjugated secondary antibodies were purchased from Cell Signaling Technology.
Western Blotting
Protein estimation of RIPA and nuclear extracts was done by the bicinchoninic acid (BCA) method, and Western blot analysis was performed using pooled protein extracts as described previously15.
Immunofluorescence Staining
Fresh-frozen liver sections (5 μm thick) were used to detect pH2AX Ser139 immunofluorescence as described previously16.
Real-Time PCR
Total RNA was isolated from APAP300 and APAP600 livers using the TRIzol method according to the manufacturer’s protocol (Sigma-Aldrich, St. Louis, MO, USA) and converted to cDNA as previously described4. Gene expression of various genes was determined by comparing mRNA levels from APAP-treated groups at different time points with 0-h control group using real-time PCR analysis. SYBR Green technology was used for real-time PCR analysis on the Applied Biosystems Prism 7300 Real-time PCR Instrument according to the manufacturer’s protocol. The 18S gene expression in the same sample was used for data normalization. Primers used for real-time PCR are listed in Table 1.
Table 1.
Primers Used in This Study
| Gene | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| GADD45α | CCGAAAGGATGGACACGGTG | TTATCGGGGTCTACGTTGAGC |
| GADD45β | CAACGCGGTTCAGAAGATGC | GGTCCACATTCATCAGTTTGGC |
| GADD45γ | GGGAAAGCACTGCACGAACT | AGCACGCAAAAGGTCACATTG |
| BTG2 | ATGAGCCACGGGAAGAGAAC | GCCCTACTGAAAACCTTGAGTC |
| PAI1 | TTCAGCCCTTGCTTGCCTC | ACACTTTTACTCCGAAGTCGGT |
| 18S | TTGACGGAAGGGCACCACCAG | GCACCACCACCCACGGAATCG |
Statistical Analysis
Data are shown as mean ± SEM. Student’s t-test was used for statistical analysis. Difference between groups was considered statistically significant at a value of p < 0.05.
RESULTS
Sustained Liver Injury and Inhibited Liver Regeneration Following Higher Dose of APAP
Liver injury after APAP300 and APAP600 treatment was assessed using serum ALT and histopathological analysis of liver tissue over a 0- to 96-h time course5. At both doses, serum ALT activity was increased and peaked at 12 h after treatment. In APAP300-treated mice, serum ALT activity regressed after 24 h and returned to normal by 72 and 96 h. However, in APAP600-treated mice, ALT activity remained higher up to 24 h and later decreased, but was persistently higher until 96 h after APAP treatment compared to APAP300 (Fig. 1A). All mice receiving APAP300 dose recovered from injury, whereas mice with APAP600 treatment showed 25% lethality, and the remaining mice had sustained injury up to 96 h5.
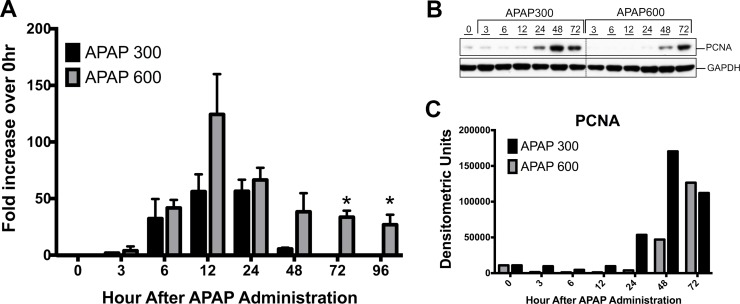
Figure 1.
Sustained liver injury and inhibited liver regeneration following a higher dose of acetaminophen (APAP). (A) Liver injury analysis by serum alanine aminotransferase (ALT) levels after APAP treatment. Shown as fold increase in ALT levels compared to 0 h. (B) Western blot analysis of proliferating cell nuclear antigen (PCNA) in whole-liver extract. (C) Densitometric analysis of PCNA Western blot. *p < 0.05 (APAP300 vs. APAP600).
To determine the difference in liver regeneration after two doses of APAP, we determined the expression of PCNA in mice liver5. Western blot analysis of PCNA revealed significantly delayed and reduced cell proliferation after the APAP600 dose compared to the APAP300 dose (Fig. 1B and C). In the APAP300 group, a significant increase in PCNA was observed from 24 h up to 72 h. However, in the APAP600 group, PCNA expression was delayed to 48 h, and it was significantly lower than the APAP300 group.
Prolonged DNA DSB and Reduced Repair Protein Expression After Higher Dose of APAP
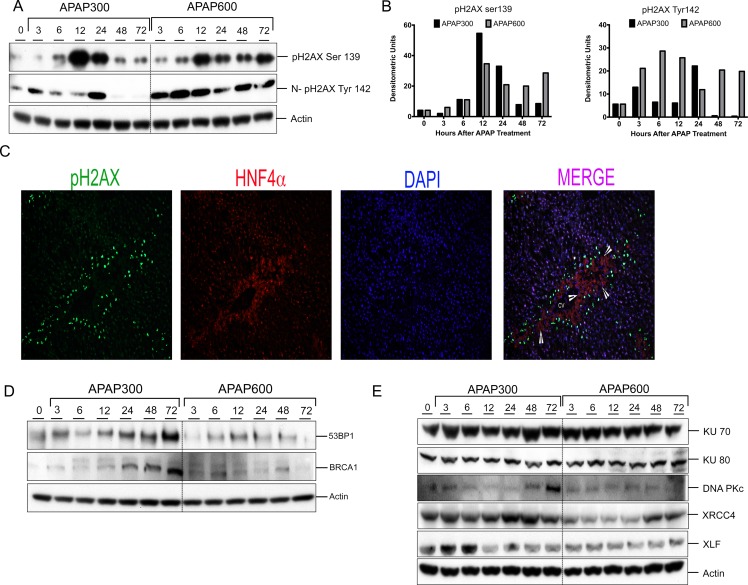
To examine the mechanism underlying delayed and attenuated cell proliferation in higher-dose mice with regard to DNA replication, we determined the most lethal form of replication stress [i.e., DNA double-strand break (DSB)]. DSB was determined using Western blot analysis and immunofluorescence detection of Ser139 phosphorylation of histone H2AX (pH2AX), a hallmark of DSB. After both APAP300 and APAP600 doses, increased pH2AX Ser139 expression was observed starting from 6 h (Fig. 2A and B). In the APAP300 group, pH2AX Ser139 expression peaked at 12 h, remained high at 24 h, and returned to control level at 48 and 72 h. In the APAP600 group, pH2AX Ser139 induction peaked at 12 h and remained high up to 72 h. Further, we studied tyrosine 142 phosphorylation (and dephosphorylation) on pH2Ax, a signal involved in the recruitment of DNA DSB repair proteins. Previous studies have shown that pH2AX dephosphorylation at Tyr142 is a critical in recruitment of repair proteins17,18. Western blot analysis of nuclear extracts showed an initial increase in pH2AX Tyr142 phosphorylation up to 24 h and a significant decrease at 48 and 72 h in the APAP300 group, indicating initiation of DNA repair. In contrast, the APAP600 group mice had higher levels of pH2AX Tyr142 throughout the time course (Fig. 2A and B). To determine which cells exhibit pH2AX after APAP overdose, we performed double immunofluorescence staining with pH2AX and hepatocyte marker HNF4α. Immunofluorescence staining revealed that hepatocytes immediately surrounding the APAP-induced necrotic zone were positive for DSB (Fig. 2C).
Figure 2.
Prolonged DNA double strand break (DSB) and reduced repair protein expression after a higher dose of APAP. (A) Western blot analysis of phos-H2AX Ser139 using total liver extract and phosphorylated histone H2AX (pH2AX) Tyr142 using nuclear extract. (B) Bar graphs showing densitometric analysis of pH2AX Ser139 and Tyr142 Western blots. (C) Representative immunofluorescence staining for pH2AX Ser139 (green), HNF4α (red), and DAPI (blue) for cell nuclei. Arrowheads are pointing to necrotic cells. (D) Western blot analysis of DNA repair mediator proteins 53BP1 and BRCA1 in total liver extract. (E) Western blot analysis of DNA repair effector proteins KU70, KU80, DNAPkc, XRCC4, XLF, and Lig4 using total liver extract.
BRCA1 and 53BP1 are critical mediator proteins involved in DDR, which can interact with broken DNA ends and help the binding of DNA repair effector proteins at the damaged DNA site19,20. A marked increase in 53BP1 and BRCA1 protein levels was seen from 12 h up to 72 h after APAP300 treatment compared to the 0-h control. In contrast, 53BP1 and BRCA1 protein expression was downregulated after APAP600 treatment (Fig. 2D).
We have previously demonstrated that in the APAP600 group, cells were arrested at the G0/G1 phase5. In the G0/G1 phase of the cell cycle, DSB is repaired, mainly by nonhomologous end joining (NHEJ)21. Therefore, we studied NHEJ repair pathway proteins to further examine the difference in DNA repair between the APAP300 and APAP600 groups. We determined the expression of proteins involved in NHEJ repair including KU70, KU80, DNA Pkc, XRCC4, XLF, and DNA Lig4 using Western blot analysis (Fig. 2E). The data indicated significant upregulation of XRCC4, XLF, DNA Pkc, and Lig4 in APAP300, all of which were downregulated in the APAP600 group compared to the 0-h control (Fig. 2E). We did not observe any difference in the KU70 and KU80 protein levels between the APAP300 and APAP600 groups.
These data suggest that after a higher dose of APAP, there is reduced DNA repair protein expression and inadequate chromatin modification resulting in impaired DSB repair.
Reduced PARP Activation Following Higher Dose of APAP
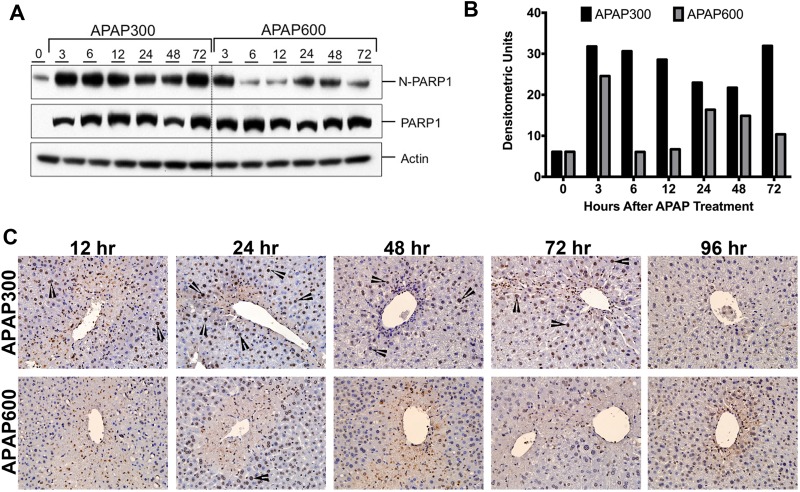
Another critical protein in DDR is poly(ADP) ribose polymerase (PARP1) that can sense the DNA damage and mediate the stress response by poly(ADP) ribosylation of nuclear proteins. It results in chromatin remodeling, which favors DNA repair22. We did not observe any difference in total PARP1 protein expression between the APAP300 and APAP600 group. However, nuclear PARP1 (N-PARP1) was significantly downregulated in the APAP600-treated mice compared to the APAP300-treated mice (Fig. 3A and B). Next, we determined PARP activation by staining for PARylated proteins using immunohistochemistry (Fig. 3C). Following the APAP300 dose, PARP activation was observed in a time-dependent manner. No PAR staining was evident until 12 h (data not shown), but significant nuclear PAR staining was evident from 12 to 72 h after APAP300 treatment. PAR staining intensity significantly increased at 24 h, was sustained until 72 h, and disappeared by 96 h after APAP300 treatment. On the contrary, in the APAP600-treated mice, at 12 and 24 h, very few cells stained positive for PAR with low intensity. At 48 h, many PAR+ cells were observed; however, staining intensity was weak compared to the APAP300 dose. PAR staining disappeared by 72 h following APAP600 treatment. These data indicate that PARP activation is significantly higher and sustained following APAP300 treatment; however, it is delayed and weak following APAP600 treatment.
Figure 3.
Delayed activation of PARP following APAP600 treatment. (A) Western blot analysis and (B) densitometric analysis of the PARP1 blots in total liver extract and nuclear extract. (C) Immunohistochemical analysis of PARylated proteins. Arrowheads point to nuclear PAR staining.
Increased Transcriptional Activation of p53 at Higher Dose of APAP
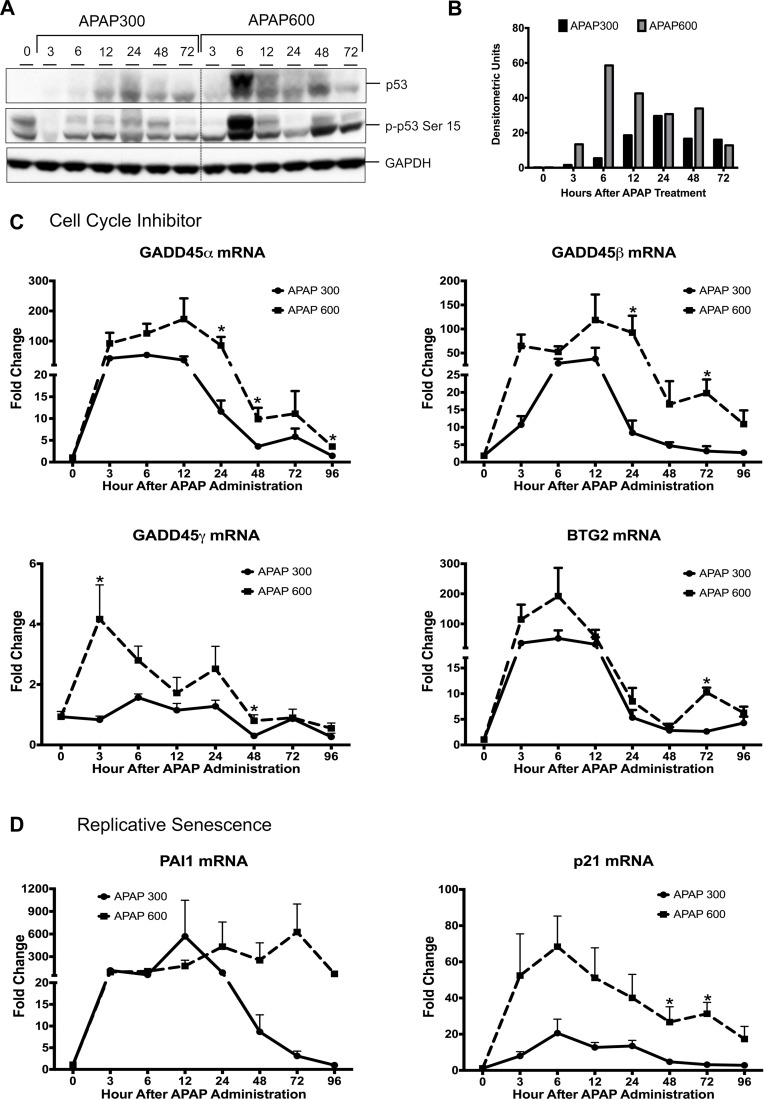
p53 is the major effector protein of the DDR pathway, which can activate cell cycle checkpoint and arrest the cell cycle until damage is repaired. Stabilization and activation of p53 protein have been shown to play an important role in many cellular processes such as cell cycle arrest, cell senescence, cell death, and cell metabolism23,24. We determined p53 activation by measuring changes in p53 protein by Western blotting and quantifying mRNA for several p53 target genes after APAP overdose. Western blot analysis of APAP300 and APAP600 samples indicated marked increase in p53 stabilization after APAP treatment (Fig. 4A and B). Interestingly, p53 protein levels were significantly higher in the APAP600 group compared to the APAP300 group from 6 h up to 72 h. APAP600-treated mice exhibited a sixfold higher p53 expression at 6 h after APAP treatment. Similarly, Ser15 phosphorylation of p53, which indicates activation of p53, was significantly higher in APAP600 at all time points (Fig. 4A). Real-time PCR analysis showed that expression of several p53-responsive genes involved in cell cycle inhibition (GADD45α, GADD45β, GADD45γ, and BTG2) (Fig. 4C) and cell senescence (PAI1) (Fig. 4D) increased consistently with increased p53 activity. Previously, we have demonstrated that p21 mRNA is significantly higher with APAP600, the nonregenerating dose, compared to APAP300, the regenerating dose5. A marked increase in all cell cycle inhibitor and cell senescence gene expressions was seen in both groups after APAP treatment. In the APAP300 group, mRNA levels of all these genes were significantly reduced from 24 to 96 h. However, these cell cycle inhibitor and senescence gene expressions were sustained and significantly higher in the APAP600 group. These data indicate that sustained activation of p53 after APAP600 treatment results in cell cycle arrest and replicative senescence.
Figure 4.
Activation of p53 is higher following APAP600 treatment. (A) Western blot analysis of total p53 and phospho-p53 Ser15 using total liver extract. (B) Bar graph showing densitometric analysis of total p53 Western blots. (C) Real-time PCR analysis of p53 target genes regulating cell cycle inhibition Gadd45α, Gadd45β, Gadd45γ, and BTG2 and (D) replicative senescence PAI1 and p21. *p < 0.05 (APAP300 vs. APAP600).
DISCUSSION
APAP is a safe analgesic and antipyretic drug when taken at the recommended daily dose. It is safely metabolized in the liver and excreted in urine. However, overdose of APAP causes ALI and even ALF, which is the number one cause of ALF in the US and UK2,25. The mechanism of APAP toxicity involves generation of ROS, release of endonucleases, extensive DNA fragmentation, and subsequent cell necrosis26. In response to injury, healthy hepatocytes surrounding the necrotic zone divide rapidly and help repair the injured liver4,5,27. In a previous study, we demonstrated that liver regeneration is stimulated rapidly following treatment with 300 mg/kg of APAP (regenerating dose), but it is significantly delayed and blunted after a 600-mg/kg dose of APAP (nonregenerating dose)5. The main reason behind this is the cells that surround the necrotic zone, which normally undergo proliferation to fuel liver regeneration, are arrested in mice treated with the higher nonregenerating dose of APAP. Our previous studies have shown that the reason behind the delayed regeneration following APAP600 treatment is not lack of enough number of viable cells. Even after this high dose, only about 40% hepatocytes at a maximum undergo necrosis, leaving up to 60% hepatocytes intact to divide and replace the dead cells. However, these cells are extremely stressed due to ongoing injury and are incapable of entering the cell cycle as shown by G0 to G1 arrest in the previous work. However, the mechanisms behind this cell cycle arrest in these stressed hepatocytes in the APAP600-treated mice are not completely known.
In the present study, we determined if this cell cycle arrest at the nonregenerating doses is due to enhanced DNA damage and blunted DNA repair processes. Our data indicate that DNA damage occurs following both the regenerating (APAP300) and nonregenerating (APAP600) doses of APAP, but the DNA repair process is significantly inhibited following treatment with the nonregenerating dose of APAP. Furthermore, immunofluorescence data revealed that the hepatocytes immediately next to the necrotic zone exhibit extensive DNA damage. These are the same hepatocytes that are required to proliferate in order to ensue liver regeneration. Whereas previous studies have shown that DNA damage is part of necrotic cell death after APAP, our data are the first to demonstrate that DNA damage and subsequent lower DNA repair inhibit liver regeneration, repair, and recovery after APAP overdose.
Because we observed sustained DSB in nonregenerating animals, we further studied whether DSB repair is inactive and cells are arrested due to failure to replicate damaged DNA with APAP600. Dephosphorylation of pH2AX at Tyr142 is one of the chromatin modifications that facilitate DSB repair17,18. We observed that Tyr142 phosphorylation was maintained for a significantly longer time following treatment with the nonregenerating APAP600 dose, which would delay recruitment of DNA repair proteins. Additionally, higher dose of APAP suppressed expression of mediator proteins 53BP1 and BRCA1 (Fig. 2D). Expression of several DSB repair effector proteins was suppressed at nonregenerating doses. Because DSB repair is the collective effort of various proteins, lack of several critical proteins will result in delayed or completely suppressed DAB repair in APAP600-treated mice. Furthermore, dephosphorylation of H2AX at Tyr142, which is required for easy access of repair proteins to DSB sites, was significantly lower in APAP600-treated mice. This may have made the damage site inaccessible for the repair protein in nonregenerating dose-treated mice. These results collectively show that DSB repair is deregulated in nonregenerating animals, leading to sustained DSB.
Further studies showed that APAP600-treated mice exhibited reduced nuclear PARP1 levels despite similar amounts of total PARP1 levels compared to APAP300. Normally, PARP1 is present in the nucleus, where it is involved in protein PARylation28,29. Our data indicate that at 6 h following APAP600, PARP1 is rapidly removed from the nucleus while PARP levels are maintained in APAP300-treated mice. This was consistent with significantly decreased PARylation of nuclear proteins in the APAP600 group. However, we did not see any significant difference in total PARP levels in either the APAP300- or APAP600-treated mice over the time course. These data indicate that following the high dose of APAP (APAP600), PARP may be actively exported out of the nucleus, reducing its nuclear activity. The mechanism of this enhanced nuclear transport is not clear. In agreement with a previous study30, these data show that PARP activation is not associated with increased liver injury following APAP toxicity. However, these data suggest that PARP activation is a critical step in DSB repair following APAP overdose. Further studies are required to delineate the mechanism of nuclear export of PARP1 following a higher dose of APAP.
p53 is a primary effector protein that plays a critical role in cell cycle regulation during DDR. Under stress conditions, p53 is stabilized and activated through various posttranslational modifications. One such modification is phosphorylation at Ser15 that leads to transcriptional activation of p53. Activated p53 regulates a plethora of downstream gene expressions involved in cell cycle inhibition and senescence24. Our data indicate significantly higher and sustained activation of p53 following the APAP600 dose (Fig. 4A). The expression of p53 target genes (cell cycle inhibitor: GADD45α, GADD45β, and GADD45γ; cell senescence: PAI1 and p21) increased after both APAP300 and APAP600, but it was significantly higher at the APAP600 dose at all time points. Previous studies indicate that moderate activation of p53 results in cell cycle arrest that permits cell to repair the DNA damage; however, excessive and sustained activation of p53 results in replicative senescence and cell death23. These data suggest that moderate activation of p53 at the regenerative dose results in transient cell cycle arrest, whereas sustained excessive activation of p53 at the nonregenerative dose may cause prolonged growth arrest and replicative senescence. Further studies are required to demonstrate the exact role of p53 and some of these target genes in liver regeneration after APAP overdose.
In conclusion, our study indicates that DNA damage and repair response plays a critical role in deciding whether liver regeneration will be “timely” or “delayed” following APAP overdose. At high doses of APAP, DSB repair is impaired, resulting in inhibited liver regeneration. This study is the first to highlight the complex signaling pathway involved in DNA DSB repair in the regulation of liver regeneration following APAP-induced ALI. These data also indicate that improving DNA repair may have therapeutic benefit after APAP overdose.
ACKNOWLEDGMENTS
This study was supported by NIH-COBRE (P20 RR021940-03), NIEHS Toxicology Training Grant (T32ES007079-34), and NIDDK R0198414.
REFERENCES
- 1. Shiffman S, Rohay JM, Battista D, Kelly JP, Malone MK, Weinstein RB, Kaufman DW. Patterns of acetaminophen medication use associated with exceeding the recommended maximum daily dose. Pharmacoepidemiol Drug Saf. 2015;24(9):915–21. [DOI] [PubMed] [Google Scholar]
- 2. Larson AM, Polson J, Fontana RJ, Davern TJ, Lalani E, Hynan LS, Reisch JS, Schiodt FV, Ostapowicz G, Shakil AO, and others. Acetaminophen-induced acute liver failure: Results of a United States multicenter, prospective study. Hepatology 2005;42(6):1364–72. [DOI] [PubMed] [Google Scholar]
- 3. Ostapowicz G, Fontana RJ, Schiodt FV, Larson A, Davern TJ, Han SH, McCashland TM, Shakil AO, Hay JE, Hynan L, and others. Results of a prospective study of acute liver failure at 17 tertiary care centers in the United States. Ann Intern Med. 2002;137(12):947–54. [DOI] [PubMed] [Google Scholar]
- 4. Apte U, Singh S, Zeng G, Cieply B, Virji MA, Wu T, Monga SP. Beta-catenin activation promotes liver regeneration after acetaminophen-induced injury. Am J Pathol. 2009;175(3):1056–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhushan B, Walesky C, Manley M, Gallagher T, Borude P, Edwards G, Monga SP, Apte U. Pro-regenerative signaling after acetaminophen-induced acute liver injury in mice identified using a novel incremental dose model. Am J Pathol. 2014;184(11):3013–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Donahower BC, McCullough SS, Hennings L, Simpson PM, Stowe CD, Saad AG, Kurten RC, Hinson JA, James LP. Human recombinant vascular endothelial growth factor reduces necrosis and enhances hepatocyte regeneration in a mouse model of acetaminophen toxicity. J Pharmacol Exp Ther. 2010;334(1):33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu B, Colletti LM. Stem cell factor and c-kit are involved in hepatic recovery after acetaminophen-induced liver injury in mice. Am J Physiol Gastrointest Liver Physiol. 2008;295(1):G45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. James LP, Lamps LW, McCullough S, Hinson JA. Interleukin 6 and hepatocyte regeneration in acetaminophen toxicity in the mouse. Biochem Biophys Res Commun. 2003;309(4):857–63. [DOI] [PubMed] [Google Scholar]
- 9. Schmidt LE, Dalhoff K. Alpha-fetoprotein is a predictor of outcome in acetaminophen-induced liver injury. Hepatology 2005;41(1):26–31. [DOI] [PubMed] [Google Scholar]
- 10. Sulli G, Di Micco R, d’Adda di Fagagna F. Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat Rev Cancer 2012;12(10):709–20. [DOI] [PubMed] [Google Scholar]
- 11. Zhou BB, Elledge SJ. The DNA damage response: Putting checkpoints in perspective. Nature 2000;408(6811):433–9. [DOI] [PubMed] [Google Scholar]
- 12. Carvajal LA, Manfredi JJ. Another fork in the road—Life or death decisions by the tumour suppressor p53. EMBO Rep. 2013;14(5):414–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ray SD, Sorge CL, Raucy JL, Corcoran GB. Early loss of large genomic DNA in vivo with accumulation of Ca2+ in the nucleus during acetaminophen-induced liver injury. Toxicol Appl Pharmacol. 1990;106(2):346–51. [DOI] [PubMed] [Google Scholar]
- 14. Shen W, Kamendulis LM, Ray SD, Corcoran GB. Acetaminophen-induced cytotoxicity in cultured mouse hepatocytes: Effects of Ca(2+)-endonuclease, DNA repair, and glutathione depletion inhibitors on DNA fragmentation and cell death. Toxicol Appl Pharmacol. 1992;112(1):32–40. [DOI] [PubMed] [Google Scholar]
- 15. Wolfe A, Thomas A, Edwards G, Jaseja R, Guo GL, Apte U. Increased activation of the Wnt/beta-catenin pathway in spontaneous hepatocellular carcinoma observed in farnesoid X receptor knockout mice. J Pharmacol Exp Ther. 2011;338(1):12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walesky C, Gunewardena S, Terwilliger EF, Edwards G, Borude P, Apte U. Hepatocyte-specific deletion of hepatocyte nuclear factor-4alpha in adult mice results in increased hepatocyte proliferation. Am J Physiol Gastrointest Liver Physiol. 2013;304(1):G26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature 2009;458(7238):591–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, Ishibe-Murakami S, Wang B, Tempst P, Hofmann K, and others. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature 2009;457(7225):57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15(1):7–18. [DOI] [PubMed] [Google Scholar]
- 20. Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis 2002;23(5):687–96. [DOI] [PubMed] [Google Scholar]
- 21. Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 2012;47(4):497–510. [DOI] [PubMed] [Google Scholar]
- 22. Krishnakumar R, Kraus WL. The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol Cell 2010;39(1):8–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell 2009;137(3):413–31. [DOI] [PubMed] [Google Scholar]
- 24. Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9(5):402–12. [DOI] [PubMed] [Google Scholar]
- 25. Bernal W, Wendon J. Acute liver failure. N Engl J Med. 2013;369(26):2525–34. [DOI] [PubMed] [Google Scholar]
- 26. Jaeschke H, Bajt ML. Intracellular signaling mechanisms of acetaminophen-induced liver cell death. Toxicol Sci. 2006;89(1):31–41. [DOI] [PubMed] [Google Scholar]
- 27. Bajt ML, Knight TR, Farhood A, Jaeschke H. Scavenging peroxynitrite with glutathione promotes regeneration and enhances survival during acetaminophen-induced liver injury in mice. J Pharmacol Exp Ther. 2003;307(1):67–73. [DOI] [PubMed] [Google Scholar]
- 28. Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): Novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7(7):517–28. [DOI] [PubMed] [Google Scholar]
- 29. Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13(7):411–24. [DOI] [PubMed] [Google Scholar]
- 30. Cover C, Fickert P, Knight TR, Fuchsbichler A, Farhood A, Trauner M, Jaeschke H. Pathophysiological role of poly(ADP-ribose) polymerase (PARP) activation during acetaminophen-induced liver cell necrosis in mice. Toxicol Sci. 2005;84(1):201–8. [DOI] [PubMed] [Google Scholar]