Abstract
Long noncoding RNAs (lncRNAs) have a variety of biological functions and play key roles in many diseases. However, the knowledge of lncRNA function during a stroke is limited. We analyzed the expression profiles of lncRNAs in the brain ischemic region of mice after a 45 min middle cerebral artery occlusion (MCAO) with a 48h reperfusion. Gene ontology and pathway analysis were used to elucidate the potential functions of the differentially expressed mRNAs. A total of 255 lncRNAs (217 up-regulated and 38 down-regulated) and 894 mRNAs (870 up-regulated and 24 down-regulated) showed significantly altered expression in the ischemic brain compared to the sham controls (fold change >2, P < 0.05). The gene ontology terms were mainly associated with neutrophil chemotaxis, positive regulation of inflammatory response, cell cycle, positive regulation of apoptotic process, and apoptotic process. The pathway analysis indicated that the mRNAs were mainly associated with inflammatory pathways. Additionally, the interactions between the differentially expressed lncRNAs and mRNAs are revealed by a dynamic lncRNA-mRNA network. Our findings provide an overview of aberrantly expressed lncRNAs in stroke and further broaden the understanding of stroke pathogenesis.
Keywords: stroke, ischemia/reperfusion, long noncoding RNA, mRNAs, bioinformatics
1. Introduction
Stroke is one of the leading causes of human death and disability worldwide. Tissue plasminogen activator (tPA) is the only pharmacological treatment that has been approved by the American Food and Drug Administration (FDA) for acute ischemic stroke[1]. In addition, mechanical devices have been approved by FDA to remove blood clot within 6 hours after stroke [2–4], and this time window is even extended 24 hours for patients with mismatch between deficit and infarct [5]. Despite these exciting progresses, only a small portion of stroke patients can receive such treatment. Therefore, it is urgent to understand the underlying mechanisms of brain injury after stroke for the purpose of exploring novel stroke treatments. The important regulatory roles of microRNAs in stroke have previously been studied by us and others [6–9], however, the functional significance and molecular mechanisms of other classes of non-coding RNAs in the regulation of ischemia/reperfusion after stroke remain unknown.
LncRNAs (long non-coding RNAs) are a unique class of RNAs longer than 200 nucleotides without evident protein coding functions [10], which have been considered to be merely transcriptional noise for a long time [11]. However, it is increasingly recognized that lncRNAs are crucial in controlling the function of the homologous mRNAs [12]. The dysregulation of these lncRNAs has been associated with many human diseases, such as cancers [11], diseases of the central nervous system [13], kidney, and cardiovascular diseases [14]. A few studies have indicated that certain lncRNAs play important roles in cerebral ischemia [15–17]. Few studies to date, however, have systematically evaluated changes in lncRNA expression after stroke. In addition, it remains unclear whether lncRNA expression profiles are related to those of protein coding mRNAs in the ischemic brain after stroke.
In this study, we aim to provide insights related with lncRNAs and mRNAs in the pathogenesis of stroke. We identified the expression profiles of both lncRNAs and mRNAs in the ischemic regions of stroke in mice, and analyzed the relationships between the expression levels of lncRNAs and mRNAs by constructing a co-expression network of lncRNAs and mRNAs.
2. Materials and methods
2.1. Focal cerebral ischemia in mice
Adult male C57BL/6 mice weighing 20–22g were purchased from Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China) and housed in temperature-controlled rooms with 12 h light/dark cycles, where they had free access to food and water. All procedures in this study were conducted according to the guidelines set by the University Animal Care and Use Committee of Capital Medical University.
Transient focal cerebral ischemia was induced by a 45-min occlusion of the right middle cerebral artery (MCA), as has been previously described [18]. In brief, anesthesia was induced by inhalation of 5% isoflurane (Lunan Pharmaceutical Group Corporation; Shandong, China) in a 30% O2, 68.5% N2O mixture, and maintained with 2% isoflurane inhalation. Rectal temperature was maintained at 37 ±0.5°C with a heating pad during surgical procedures. Sham-operated animals underwent sham surgery under anesthesia, but without the right MCA occlusion. Mice were placed in a post-operative cage, and kept warm and undisturbed for a minimum of 2 h for observation. The mouse brains were removed 48 h after reperfusion. The ischemic regions were collected and immediately frozen in liquid nitrogen cans.
2.2. Measurement of ischemic infarct sizes
Mice were sacrificed 48 h after MCAO, and the brain was quickly removed and cut into 1 mm thick coronal sections. The sections were immersed in 2% TTC at 37 °C for 20 min and then fixed in 4% paraformaldehyde. The infarct size of the ischemic cortex was normalized to the non-ischemic cortex and expressed as a percentage [19].
2.3. LncRNA microarray and analysis
Total RNA containing small RNA was extracted using TRIzol reagent (Invitrogen, Canada) according to the manufacturer’s protocol. RNA purity and concentration were determined with NanoDrop ND-1000 spectrophotometry, and the RNA integrity was determined via 1% gel electrophoresis.
The microarray analysis was performed with the Affymetrix GeneChip Mouse Transcriptome Array 1.0, which was designed with about 55,000 mouse lncRNAs and 23,000 mouse genes. The purified RNA was transcribed into complementary DNA (cDNA) according to the protocol set forth by Gminix (Shanghai, PR China). The cDNA was fragmented, labeled with fluorescent dyes and was then incubated in the GeneChip Hybridization Oven 645. After washing the chip, the GeneChip Scanner 3000 7G was used to measure the fluorescence intensity. After image data were saved, the probe summarization was performed using the software Expression Console (version 1.2.1).
We used the significance analysis of microarrays (SAM) model to identify the differentially expressed lncRNAs and mRNAs between the ischemic and sham groups. After a significant analysis and false-discovery rate (FDR) analysis, a P value <0.05 was regarded as significant. Moreover, fold changes greater than 2 or less than 0.5 were considered to be higher fold changes.
2.4. Quantitative real-time PCR (qRT-PCR) assay
Total RNA was isolated using the Trizol reagent and reverse transcribed into cDNA using the Thermo First cDNA Synthesis Kit (SinoGene). The qRT-PCR was conducted to test the expression of four lncRNAs using the 2×SG Green qRT-PCR Mix. All of the reagents were obtained from SinoGene. The expression levels were analyzed based on the cycle threshold (CT) values and normalized to internal β-actin. The primer sequences are listed in Table 1.
Table 1.
Primers used for RT-qPCR analysis.
| Primer name | Primer sequence (5′ -3′) forward | Primer sequence (5′ -3′) reverse |
|---|---|---|
| ENSMUST00000121456 | GGCAAGAACACCATGATGCA | ACAGCATGTCCCGAATCTCA |
| NONMMUT036055 | ATGTCGGGTTCTTCTAGCGT | TCAGCAGAGGGTCATGTTGA |
| KnowTID_00002053 | AGACTCCTCTACCCTGTGCT | AAAGCAGCGACATGAAACA |
| NONMMUT038744 | GGGGACTCGAGATCTACTGC | ATAGCTGAACTGGGCGATCA |
| β–actin | CGTTGACATCCGTAAAGACC | CTAGGAGCCAGAGCAGTAATC |
2.5. Gene ontology (GO) and kyoto Encyclopedia of genes and genomes (KEGG) analysis
GO analysis was used to investigate the roles of all differentially expressed mRNAs (http://www.geneontology.org). A pathway analysis was used to determine the significant pathways related with the differentially expressed mRNAs, according to KEGG (http://www.genome.jp/kegg/). We used Fisher’s exact test and the χ2 test to select the significant GO categories and pathways. The threshold of significance was a P value <0.05, and false discovery rate (FDR) was calculated to correct the P value.
2.6. Construction of the lncRNA-mRNA co-expression network
The mRNA-lncRNA co-expression network, which was used to identify interactions between the differentially expressed lncRNAs and mRNAs, was constructed based on Pearson’s correlation analysis. For each pair of genes, correlation coefficients of 0.95 or greater were selected to construct the network through the software Cytoscape. In the network, each gene/lncRNA corresponds to a node, and the nodes are connected by an edge. The more adjacent genes or lncRNAs to which a gene is connected, the larger its degree and the more important it is.
3. Results
3.1. Overview of lncRNA and coding gene profiles in the brains of mice following MCAO
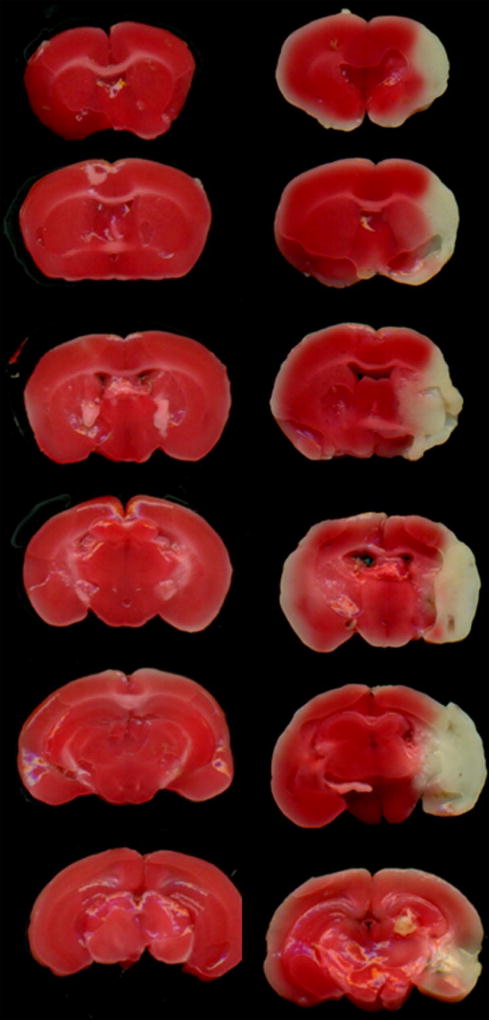
The TTC staining results showed a large area of infarction in the cerebral cortex and striatum in the ischemic group (Fig. 1). The ischemicregions corresponding to the white areas in the additional ischemic brains were dissected for the lncRNA microarray assay.
Fig 1. Ischemia/reperfusion-induced cerebral injuries in mice.
Representative TTC images from the sham and ischemia groups, n=6 per group.
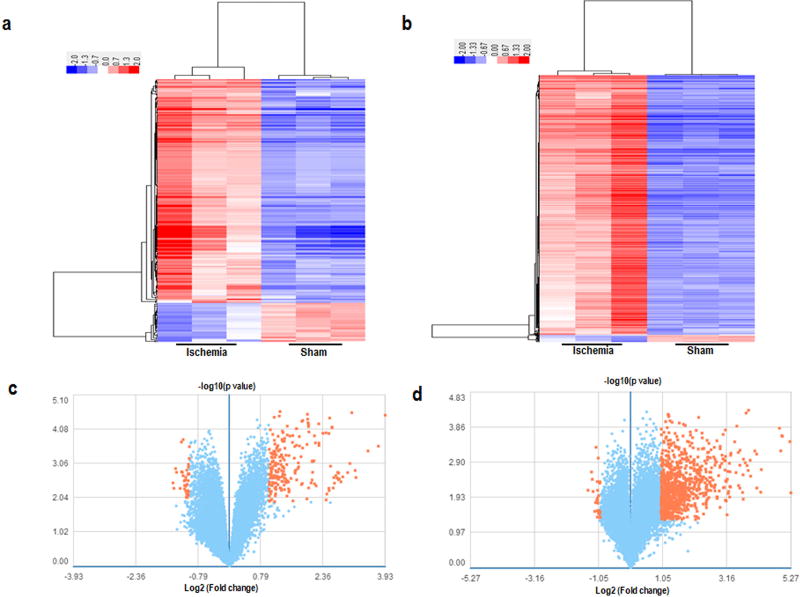
In order to characterize the lncRNA expression profile in the ischemic brains, we performed a lncRNA and mRNA microarray assay to determine their respective expression levels. The results showed that the expression of a total of 255 lncRNAs changed dramatically with at least 2 or 0.5-fold changes. Among the 255 differentially expressed lncRNAs, 217 were up-regulated, while 38 were down-regulated in the ischemic regions, compared with those in the corresponding non-ischemic brains of the sham group (Fig. 2).
Fig 2. The expression profiles of lncRNAs and mRNAs in mice from the sham and ischemic groups by microarray analysis.
The top row: the hierarchical clusterings show a remarkably differential expression of lncRNAs (a) and mRNAs (b), n=3 per group. Red and green colors represent high and low expression levels, respectively. Each column represents a single group, and each row represents an lncRNA or mRNA. The bottom row: the volcano plots of expression profiles of differentially expressed lncRNAs (c) and mRNAs (d) in mice from the sham and ischemic groups. Orange dots represent genes that passed the statistical and fold-change cutoffs.
3.2. Validation of deregulated lncRNAs in ischemic brains
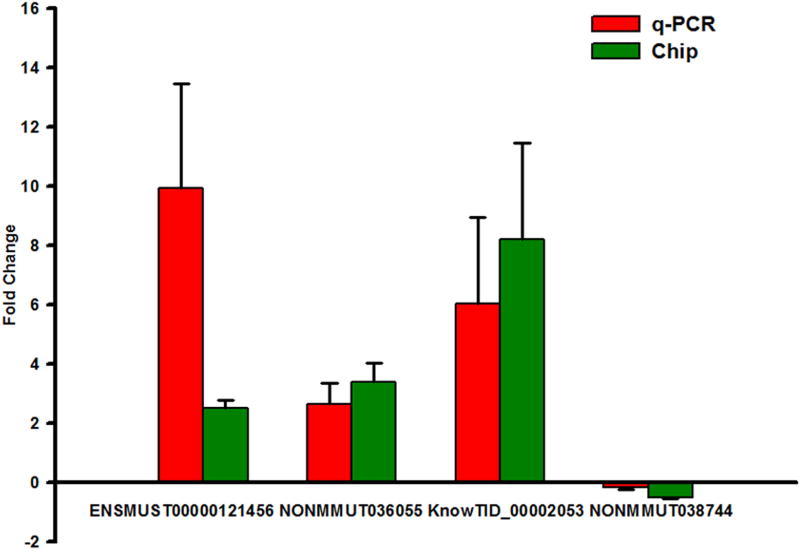
Four lncRNAs were randomly selected from the microarray analysis data for validation by using quantitative RT-PCR. As shown in Fig. 3, the qRT-PCR analysis revealed that the expressions of the lncRNAs ENSMUST00000121456, NONMMUT036055, and KnowTID_00002053 were up-regulated, whereas NONMMUT038744 expression was down-regulated in the ischemic brains (Fig. 3). These results are consistent with the microarray data.
Fig 3. Validation of the reliability of microarray data via quantitative real-time PCR.
The fold-changes represent ratios by comparing the expression values of the ischemia to sham results, as detected by microarray or qRT-PCR, n=3 per group. The positive refers to upregulation, and the negative refers to downregulation.
3.3. GO and pathway analyses of differentially expressed mRNAs
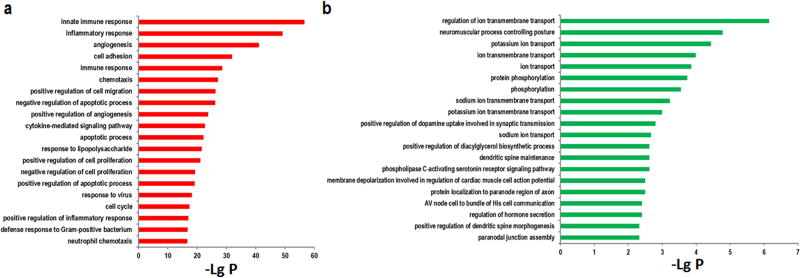
GO analysis was performed to determine the function of the differentially expressed mRNAs after a stroke. As shown in Fig. 4a and 4b, the highest enriched GOs of upregulated mRNAs were those involved in neutrophil chemotaxis, the inflammatory response, the cell cycle, and apoptosis. In contrast, the highest enriched GOs of downregulated mRNAs were those regulating ion transmembrane transport, potassium ion transport, ion transmembrane transport, and protein phosphorylation.
Fig 4. GO analysis of differentially expressed mRNAs in ischemia mice.
(a) The top 20 GO terms associated with upregulated mRNAs. (b) The top 20 GO terms associated with downregulated mRNAs. The value of −lg (p value) was calculated to reflect the significance of GO terms.
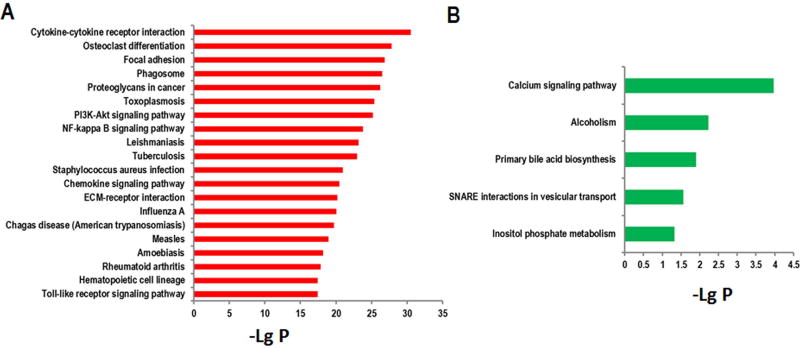
After identifying the differentially expressed mRNAs, we further performed a pathway analysis according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The results indicate that the upregulated mRNAs are involved in the regulation of a cytokine-cytokine receptor interaction, the NF-kappa B signaling pathway, the PI3K-Akt signaling pathway, and the Toll-like receptor signaling pathway (Fig. 5a). The downregulated mRNAs are involved in inositol phosphate metabolism and the Calcium signaling pathway (Fig. 5b).
Fig 5. The KEGG analysis of differentially expressed mRNAs in ischemic brains.
(a) The top 20 pathway terms associated with upregulated mRNAs. (b) The pathway terms associated with downregulated mRNAs. The value of −lg (p value) was calculated to reflect the significance of pathway terms.
3.4. Construction of the functional lncRNA–mRNA network
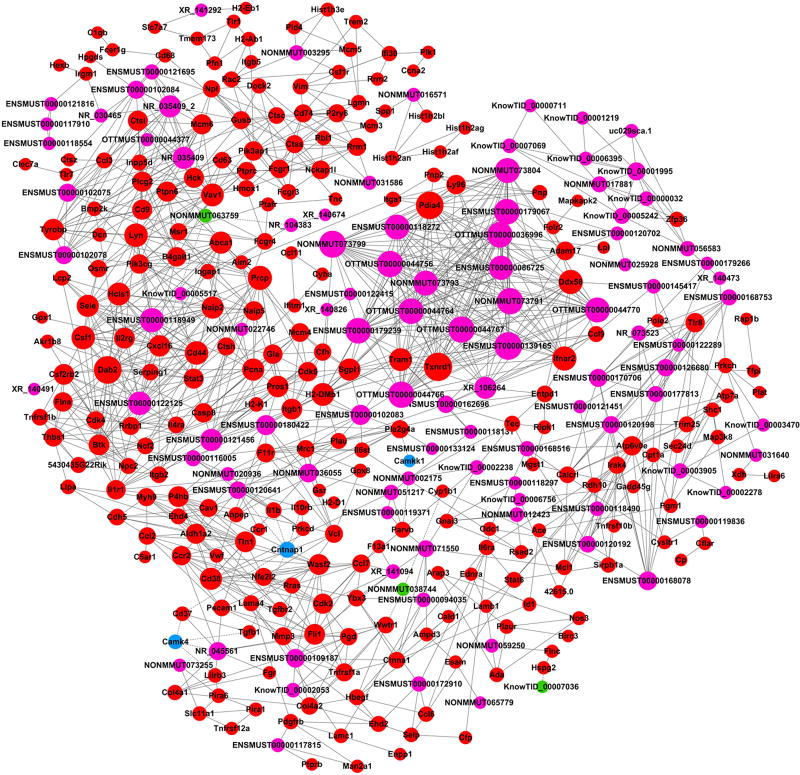
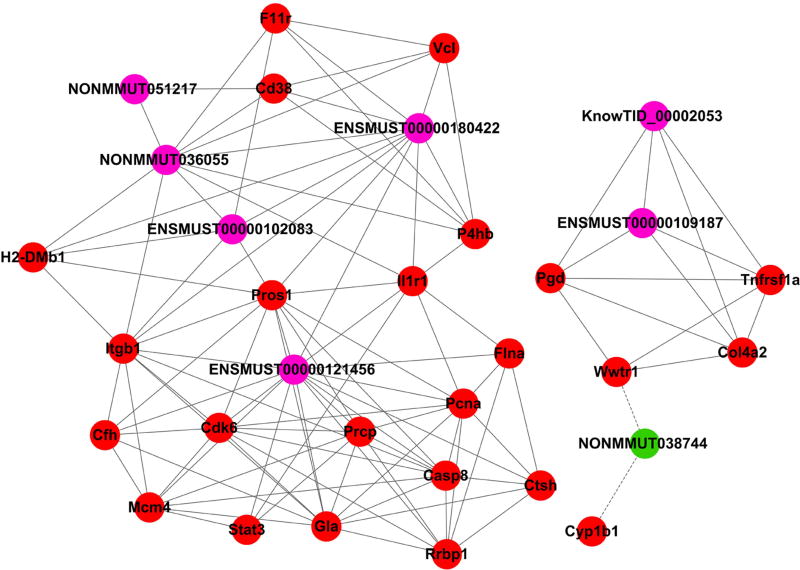
Based on the correlations of the differentially expressed lncRNAs and mRNAs, mRNA-lncRNA co-expression networks were constructed to discover the significant molecular lncRNA mechanisms in stroke (Fig. 6). The co-expression network shows that one lncRNA was associated with one to dozens of mRNAs, suggesting that there is an inter-regulation of lncRNAs and mRNAs in stroke. We further focused on co-expression networks centering on the four confirmed lncRNAs, ENSMUST00000121456, NONMMUT036055, KnowTID_00002053, and NONMMUT038744 (Fig. 7). For example, ENSMUST00000121456 expression is positively correlated with Casp 8 and Stat 3 mRNA expression levels, which are important for neuronal death, neuroinflammation, cell proliferation, and differentiation. The network suggests that lncRNA expression may regulate the corresponding mRNA expression levels to simultaneously participate in ischemic injures.
Fig 6. Co-expression networks of all differentially expressed lncRNA-mRNA in the ischemic brains.
The lncRNAs are shown in purple (upregulated) and green (downregulated), while mRNAs are presented in red (upregulated) and blue (downregulated).
Fig 7. Co-expression networks of the verified lncRNA-mRNA in ischemia mice.
The lncRNA-mRNA network containing the four verified aberrantly expressed lncRNAs. The lncRNAs are shown in purple (upregulated) and green (downregulated), while mRNAs are presented in red (upregulated).
4. Discussion
In the present study, we provide novel data showing that 255 lncRNAs and 894 mRNAs were differentially expressed in ischemic brains, and that those differentially expressed mRNAs are mainly involved in the processes of neutrophil chemotaxis, the inflammatory response, the cell cycle, and apoptosis. The KEGG pathway analysis indicates that these mRNAs are involved in the regulation of cytokine-cytokine receptor interactions, the NF-kappa B signaling pathway, and the PI3K-Akt signaling pathway. Since our lncRNA-mRNA network analyses suggest that lncRNA alterations are closely related with mRNA changes, we provide strong evidence that altering lncRNA expression might affect brain injury after stroke via regulating the expression of these differentially mRNA, which are involved in inflammation, apoptosis, and cell survival pathways.
Most previous studies have been devoted to exploring the role of small ncRNAs such as microRNAs (~21–25 nucleotides) [20]. With the development of lncRNA microarray techniques, thousands of lncRNAs have been identified. The dysregulation of lncRNAs has been associated with many human diseases, including different types of cancers and diseases of the central nervous system [21, 22]. Nevertheless, research on lncRNAs related to cerebral ischemic disease is limited.
A recent microarray profiling study showed that stroke significantly influenced the cerebral lncRNAome in rats [23]. Bioinformatic analysis has shown that these stroke-responsive lncRNAs have >90% sequence homology with protein-coding genes. In addition, stroke-induced lncRNAs were suggested to be associated with the chromatin-modifying protein Sin3A and corepressors of the RE-1 silencing transcription factor, in order to modulate the post-ischemic epigenetic landscape [24]. In addition, Zhao, et al., demonstrated that hypoxic-ischemic injury altered lncRNA expression profiles in the neonatal rat brain [25]. In particular, the lncRNA BC088414 was upregulated after hypoxic-ischemic injury and its knockdown attenuated cell apoptosis and promoted cell proliferation by reducing the mRNA levels of Casp6 and Adrb2 in PC12 cells subjected to oxygen and glucose deprivation. Few studies to date, however, have evaluated lncRNA changes in the mouse brain after ischemia-reperfusion injury, and none have systematically studied the relationship between lncRNA and mRNA expression levels using microarray techniques.
In the current study, most of these lncRNAs identified by us have not yet been functionally characterized, while most of the identified mRNAs are well known. Therefore, a bioinformatic analysis of the aberrantly expressed mRNAs was used to gain a better understanding of the putative function of the differentially expressed lncRNAs.
First, GO enrichment analyses were used to analyze the differentially expressed protein-coding genes associated with the lncRNAs. The results revealed that the upregulated genes were associated with neutrophil chemotaxis, positive regulation of inflammatory response, the cell cycle, positive regulation of the apoptotic process, and the apoptotic process. The highest enriched GO terms for downregulated mRNAs were associated with the regulation of ion transmembrane transport, potassium ion transport, ion transmembrane transport, and protein phosphorylation.
Second, the pathway analysis was conducted by KEGG database.. It is not surprising that the inflammatory response was found to hold an important position in these genes, as it is well known that the immune and inflammatory responses are major contributors to acute ischemic brain injuries [26, 27]. The NF-kappa B signaling pathway has been reported to play a vital role in the inflammation response after ischemic stroke-induced brain injuries [28, 29]. In addition, the PI3K/Akt pathway has been proven to regulate neuronal survival and also participates in the inflammatory response [30]. Furthermore, the toll-like receptor has also been demonstrated to contribute to the development and progression of inflammatory and autoimmune diseases [31]. Our results suggest that these pathways may harbor significance and/or may contribute to the pathogenesis and biochemical characteristics of ischemia/reperfusion-induced cerebral injuries in mice.
Third, we constructed a co-expression network by combining differentially expressed lncRNAs with differentially expressed mRNAs, based on their locational distributions and sequence correlations. The results suggested that the differentially expressed lncRNAs interact with the differentially expressed genes. Our novel results provide strong evidence that lncRNAs may participate in brain injury by altering mRNA expression levels.
Finally, the co-expression networks centering on four lncRNAs: suggested that ENSMUST00000121456 expression is positively correlated with Casp 8 and Stat 3 mRNA expression levels, which are important for neuronal death, neuroinflammation, cell proliferation, and differentiation. In addition, another pair with a positive correlation was NONMMUT036055 with Il1r1mRNA, which is interleukin 1 receptor, type I. Previous studies have suggested that antagonism at the IL-1R induces neuroprotection in several rodent models of neuronal injury [32]. Nevertheless, NONMMUT038744 expression was negatively correlated with Wwtr1 mRNA expression. WWTR1, also knowns as TAZ, is a transcriptional coactivator. Hepatocyte TAZ/WWTR1 has been indicated to promote inflammation and fibrosis in nonalcoholic steatohepatitis [33].
We are aware of some limitations of the current study. For example, given our research methods, we merely predicted the function of differentially expressed lncRNAs and were unable to determine exactly how these lncRNAs regulate the target gene expression. In addition, we used whole brain tissues, and thus the functions of lncRNAs in specific cells types cannot be identified. Furthermore, the brain tissues were collected 48 hours after stroke, and the time course of the expression of different genes was not studied. In the future, the functions of lncRNAs in different cell types should be studied, and their expression patterns at multiple time points should be identified. Furthermore, the functions of some key lncRNAs can be validated by modulating their expression levels via overexpression or inhibition.
In summary, we provide novel evidence that lncRNAs are differentially expressed in the ischemic brain after stroke, and that alteration to this expression is closely related with changes in gene expression, which are involved in neuroinflammation, apoptosis, cell differentiation, and proliferation. We conclude that lncRNAs may regulate brain injury by altering the expressions of associated genes and proteins.
HIGHLIGHTS.
A total of 255 lncRNAs and 894 mRNAs showed significantly altered expression in the ischemic brain.
Gene ontology and pathway analysis were used to elucidate the potential functions of the differentially expressed mRNAs.
A dynamic lncRNA-mRNA network revealed the interactions between the differentially expressed lncRNAs and mRNAs.
Acknowledgments
This work is supported by the National Natural Science Foundation of China(81701154), Beijing Natural Science Foundation (7172109) and R01NS06413606 (HZ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: none declared.
References
- 1.Wang Y, Li Q, Wang J, Zhuang QK, Zhang YY. Combination of thrombolytic therapy and neuroprotective therapy in acute ischemic stroke: is it important? European review for medical and pharmacological sciences. 2015;19(3):416–422. [PubMed] [Google Scholar]
- 2.Campbell BC, Mitchell PJ, Kleinig TJ, Dewey HM, Churilov L, Yassi N, Yan B, Dowling RJ, Parsons MW, Oxley TJ, et al. Endovascular therapy for ischemic stroke with perfusion-imaging selection. The New England journal of medicine. 2015;372(11):1009–1018. doi: 10.1056/NEJMoa1414792. [DOI] [PubMed] [Google Scholar]
- 3.Cobb MIH, Laarakker AS, Gonzalez LF, Smith TP, Hauck EF, Zomorodi AR. Endovascular Therapies for Acute Ischemic Stroke in Children. Stroke. 2017;48(7):2026–2030. doi: 10.1161/STROKEAHA.117.016887. [DOI] [PubMed] [Google Scholar]
- 4.Saver JL, Goyal M, Bonafe A, Diener HC, Levy EI, Pereira VM, Albers GW, Cognard C, Cohen DJ, Hacke W, et al. Stent-retriever thrombectomy after intravenous t-PA vs. t-PA alone in stroke. The New England journal of medicine. 2015;372(24):2285–2295. doi: 10.1056/NEJMoa1415061. [DOI] [PubMed] [Google Scholar]
- 5.Nogueira RG, Jadhav AP, Haussen DC, Bonafe A, Budzik RF, Bhuva P, Yavagal DR, Ribo M, Cognard C, Hanel RA, et al. Thrombectomy 6 to 24 Hours after Stroke with a Mismatch between Deficit and Infarct. The New England journal of medicine. 2017 doi: 10.1056/NEJMoa1706442. [DOI] [PubMed] [Google Scholar]
- 6.Dharap A, Bowen K, Place R, Li LC, Vemuganti R. Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2009;29(4):675–687. doi: 10.1038/jcbfm.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39(3):959–966. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- 8.Liu C, Peng Z, Zhang N, Yu L, Han S, Li D, Li J. Identification of differentially expressed microRNAs and their PKC-isoform specific gene network prediction during hypoxic pre-conditioning and focal cerebral ischemia of mice. Journal of neurochemistry. 2012;120(5):830–841. doi: 10.1111/j.1471-4159.2011.07624.x. [DOI] [PubMed] [Google Scholar]
- 9.Liu C, Zhao L, Han S, Li J, Li D. Identification and Functional Analysis of MicroRNAs in Mice following Focal Cerebral Ischemia Injury. International journal of molecular sciences. 2015;16(10):24302–24318. doi: 10.3390/ijms161024302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. The Journal of clinical investigation. 2016;126(8):2775–2782. doi: 10.1172/JCI84421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vemuganti R. All's well that transcribes well: non-coding RNAs and post-stroke brain damage. Neurochemistry international. 2013;63(5):438–449. doi: 10.1016/j.neuint.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng SY, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends in genetics : TIG. 2013;29(8):461–468. doi: 10.1016/j.tig.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Lorenzen JM, Thum T. Long noncoding RNAs in kidney and cardiovascular diseases. Nature reviews Nephrology. 2016;12(6):360–373. doi: 10.1038/nrneph.2016.51. [DOI] [PubMed] [Google Scholar]
- 15.Yan H, Yuan J, Gao L, Rao J, Hu J. Long noncoding RNA MEG3 activation of p53 mediates ischemic neuronal death in stroke. Neuroscience. 2016;337:191–199. doi: 10.1016/j.neuroscience.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 16.Xu Q, Deng F, Xing Z, Wu Z, Cen B, Xu S, Zhao Z, Nepomuceno R, Bhuiyan MI, Sun D, et al. Long non-coding RNA C2dat1 regulates CaMKIIdelta expression to promote neuronal survival through the NF-kappaB signaling pathway following cerebral ischemia. Cell death & disease. 2016;7:e2173. doi: 10.1038/cddis.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Li Q, Zhang KS, Hu B, Niu X, Zhou SM, Li SG, Luo YP, Wang Y, Deng ZF. Downregulation of the Long Non-Coding RNA Meg3 Promotes Angiogenesis After Ischemic Brain Injury by Activating Notch Signaling. Molecular neurobiology. 2016 doi: 10.1007/s12035-016-0270-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao H, Tao Z, Wang R, Liu P, Yan F, Li J, Zhang C, Ji X, Luo Y. MicroRNA-23a-3p attenuates oxidative stress injury in a mouse model of focal cerebral ischemia-reperfusion. Brain research. 2014;1592:65–72. doi: 10.1016/j.brainres.2014.09.055. [DOI] [PubMed] [Google Scholar]
- 19.Ren C, Gao X, Steinberg GK, Zhao H. Limb remote-preconditioning protects against focal ischemia in rats and contradicts the dogma of therapeutic time windows for preconditioning. Neuroscience. 2008;151(4):1099–1103. doi: 10.1016/j.neuroscience.2007.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipkova J, Duris K. The role of microRNA in ischemic and hemorrhagic stroke. Current drug delivery. 2016 doi: 10.2174/1567201813666160919142212. [DOI] [PubMed] [Google Scholar]
- 21.Pastori C, Wahlestedt C. Involvement of long noncoding RNAs in diseases affecting the central nervous system. RNA biology. 2012;9(6):860–870. doi: 10.4161/rna.20482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2013;34(2):613–620. doi: 10.1007/s13277-013-0658-6. [DOI] [PubMed] [Google Scholar]
- 23.Dharap A, Nakka VP, Vemuganti R. Effect of focal ischemia on long noncoding RNAs. Stroke. 2012;43(10):2800–2802. doi: 10.1161/STROKEAHA.112.669465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dharap A, Pokrzywa C, Vemuganti R. Increased binding of stroke-induced long non-coding RNAs to the transcriptional corepressors Sin3A and coREST. ASN neuro. 2013;5(4):283–289. doi: 10.1042/AN20130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao F, Qu Y, Liu J, Liu H, Zhang L, Feng Y, Wang H, Gan J, Lu R, Mu D. Microarray Profiling and Co-Expression Network Analysis of LncRNAs and mRNAs in Neonatal Rats Following Hypoxic-ischemic Brain Damage. Scientific reports. 2015;5:13850. doi: 10.1038/srep13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papp V, Molnar T, Banati M, Illes Z. Immune responses and neuroimmune modulation in the pathogenesis of acute ischemic stroke and poststroke infections. Ideggyogyaszati szemle. 2010;63(7–8):232–246. [PubMed] [Google Scholar]
- 27.An C, Shi Y, Li P, Hu X, Gan Y, Stetler RA, Leak RK, Gao Y, Sun BL, Zheng P, et al. Molecular dialogs between the ischemic brain and the peripheral immune system: dualistic roles in injury and repair. Progress in neurobiology. 2014;115:6–24. doi: 10.1016/j.pneurobio.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simmons LJ, Surles-Zeigler MC, Li Y, Ford GD, Newman GD, Ford BD. Regulation of inflammatory responses by neuregulin-1 in brain ischemia and microglial cells in vitro involves the NF-kappa B pathway. Journal of neuroinflammation. 2016;13(1):237. doi: 10.1186/s12974-016-0703-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu W, Wang X, Zheng Y, Shang G, Huang J, Tao J, Chen L. Electroacupuncture inhibits inflammatory injury by targeting the miR-9-mediated NF-kappaB signaling pathway following ischemic stroke. Molecular medicine reports. 2016;13(2):1618–1626. doi: 10.3892/mmr.2015.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams DL, Ozment-Skelton T, Li C. Modulation of the phosphoinositide 3-kinase signaling pathway alters host response to sepsis, inflammation, and ischemia/reperfusion injury. Shock (Augusta, Ga) 2006;25(5):432–439. doi: 10.1097/01.shk.0000209542.76305.55. [DOI] [PubMed] [Google Scholar]
- 31.Leifer CA, Medvedev AE. Molecular mechanisms of regulation of Toll-like receptor signaling. Journal of leukocyte biology. 2016;100(5):927–941. doi: 10.1189/jlb.2MR0316-117RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helmy A, Guilfoyle MR, Carpenter KL, Pickard JD, Menon DK, Hutchinson PJ. Recombinant human interleukin-1 receptor antagonist in severe traumatic brain injury: a phase II randomized control trial. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2014;34(5):845–851. doi: 10.1038/jcbfm.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Zheng Z, Caviglia JM, Corey KE, Herfel TM, Cai B, Masia R, Chung RT, Lefkowitch JH, Schwabe RF, et al. Hepatocyte TAZ/WWTR1 Promotes Inflammation and Fibrosis in Nonalcoholic Steatohepatitis. Cell metabolism. 2016;24(6):848–862. doi: 10.1016/j.cmet.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]