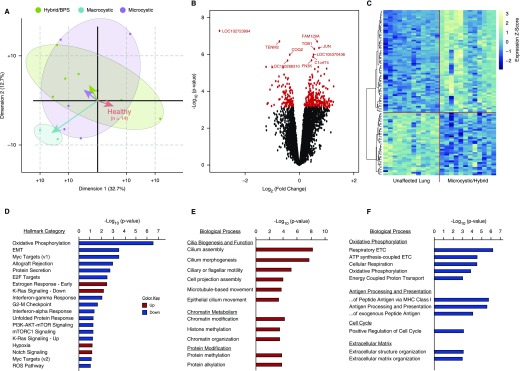
Figure 1.
The congenital lung lesion transcriptome. (A) A biplot of principal component analysis of gene expression data from 28 paired samples (from 14 patients: 6 microcystic, 5 hybrid, 1 bronchopulmonary sequestration [BPS], and 2 macrocystic lesions) demonstrates that microcystic and hybrid/BPS lesions cluster together, separate from macrocystic lesions, and from paired unaffected lung. Comparison of microcystic/hybrid/BPS lesions with paired healthy lung was used for subsequent analyses. (B) Three hundred and fifty-one genes were differentially expressed (false discovery rate, <5%) between microcystic and hybrid lesions compared with paired control lung. (C) Unbiased hierarchal clustering based on the top 100 differentially expressed genes demonstrated clear separation of the microcystic congenital pulmonary airway malformation and hybrid/BPS lesion samples from paired normal tissue. Scale to right of heat map illustrates z-score color range. (D) Gene-set enrichment analysis of differentially expressed genes between microcystic/hybrid/BPS lesions and paired unaffected lung. (E and F) Gene ontology analysis of differentially expressed genes demonstrates increased expression of genes related to airway epithelium and chromatin/histone modifications, and dysregulated expression of genes related to KRAS and PI3K–AKT–mTOR (phosphatidylinositol 3-kinase–AKT–mammalian target of rapamycin) signaling, and Myc transcriptional targets. EMT = epithelial–mesenchymal transition; ETC = electron transport chain; MHC = major histocompatibility complex; mTORC1 = mTOR complex 1; ROS = reactive oxygen species.