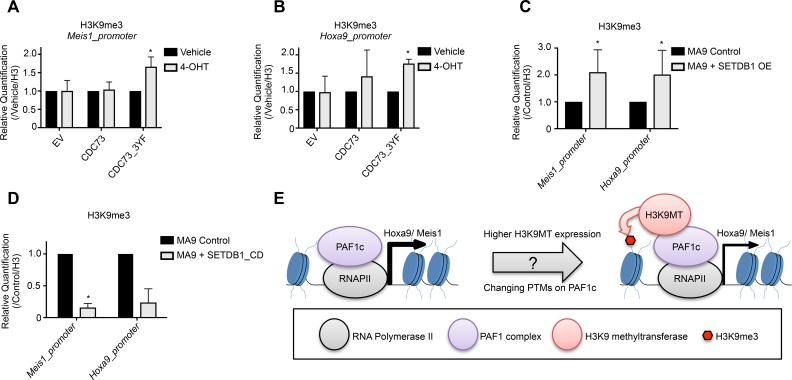
Figure 5. SETDB1 or CDC73_3YF contribute to epigenetic remodeling at the Meis1 and Hoxa9 promoter.
(A-B) ChIP-qPCR experiments for H3K9me3 at the (A) Meis1 and (B) Hoxa9 promoter were performed in MLL-AF9 transformed CDC73fl/fl-CreERT2 CDC73 re-expression cells treated with 4-OHT or vehicle control (biological replicates n=3; 2-3 technical replicates each). (C-D) ChIP-qPCR experiments for H3K9me3 at the Meis1 and Hoxa9 promoters were performed in MLL-AF9 control cells with and without transduced overexpressed (C) SETDB1 or (D) SETDB1_CD (biological replicates n=2-3; 2-3 technical replicates each). Statistical analysis was performed as described in the methods using a linear regression model with ANOVA found in Supplementary Figure 3. (E) Working model for the modulation of Hoxa9 and Meis1 transcription by the PAF1c-SETDB1 interaction. Increased expression of H3K9 methyltransferases or stabilized interaction between H3K9 methyltransferases and the PAF1c, possibly mediated by post-translational modifications (PTMs), leads to promoter H3K9 methylation and reduced expression of Hox genes. *p<0.05; EV = Empty Vector control (MigR1); OE = Overexpression.