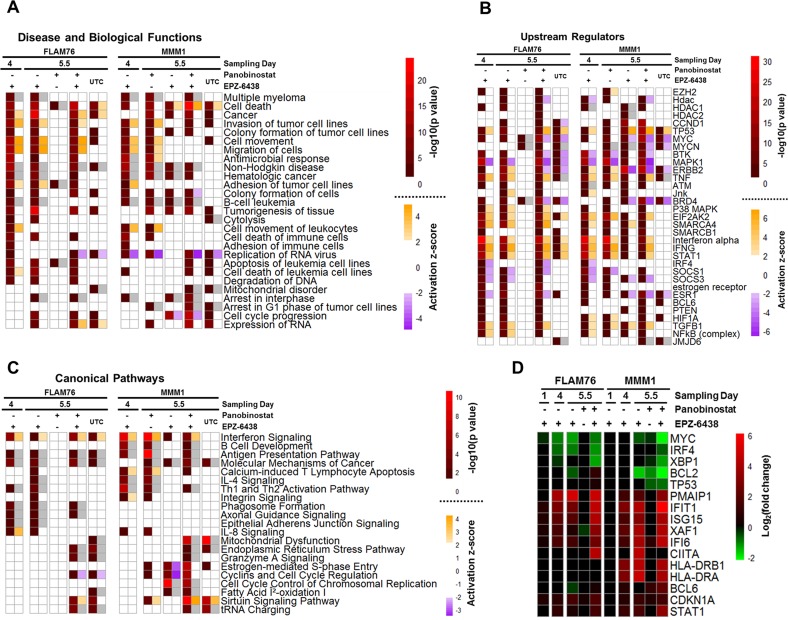
Figure 4. Ingenuity pathway analysis of EPZ-6438-, panobinostat-, and combination-induced gene expression changes.
Filtered differential gene expression profiles (FPKM≥1, |FC|≥2,FDR<0.05) for two HMCLs (FLAM76 & MMM1) treated with EPZ-6438 (μM), panobinostat (FLAM76-3 nM;MMM1-20 nM) or the combination (relative to time-matched untreated control) were subjected to Ingenuity Pathway Analysis (IPA). Heat maps represent selected top results from different types of IPA analysis: (A) predicted upstream regulators (genes, groups or complexes), (B) enriched disease and biological functions and (C) enriched canonical pathways. Each differential expression condition is represented by two columns. The left column displays the –log10(p-value) returned by IPA for enrichment and the right column displays the predicted activation z-score (when applicable; not all predictions have activation directionality). Analysis of unique-to-combination (UTC) genes subsetted from the combination condition for each line is also displayed. White heat map cells represent a lack of significant enrichment/prediction (p > 0.05 and |z|<2 respectively). Grey heat map cells represent missing activation z-scores when p-values are significant. (D) differential expression (RNA-seq: log2 fold change) across all conditions for selected genes pertinent to top IPA hits and discussed in text. Only gene expression changes significant at FDR < 0.05 and |fc|>1.5 are displayed.