Figure 1.
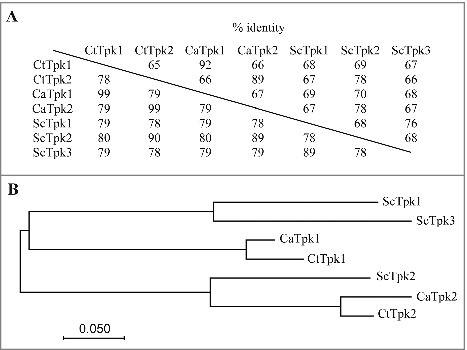
Identification of C. tropicalis PKA catalytic subunits (Tpk1 and Tpk2). (A) The amino acid identity of Tpk proteins from C. albicans (Ca), C. tropicalis (Ct), and S. cerevisiae (Sc). Numbers above the diagonal line indicate sequence identity between the full-length proteins, while numbers below represent the identity of PKA domain (accesion:cd05580). (B) Phylogenetic tree of the PKA catalytic subunits of C. albicans, C. tropicalis, and S. cerevisiae. The amino acid identity and the multiple alignment were analyzed and constructed using the Clustal Omega program (http://www.ebi.ac.uk/Tools/msa/clustalo/). The phylogenetic tree was constructed in MEGA7 [68] software using Maximum Likelihood method based on the JTT matrix-based model [69]. The black bar indicates an evolutionary distance of 0.05 substitutions per site. Proteins and GenBank accession numbers: CaTpk1, XP_723574.1; CaTpk2, XP_714866.2; CtTpk1, XP_002549018.1; CtTpk2, XP_002547429.1; ScTpk1, NP_012371.2; ScTpk2, NP_015121.1; ScTpk3, NP_012755.1.
