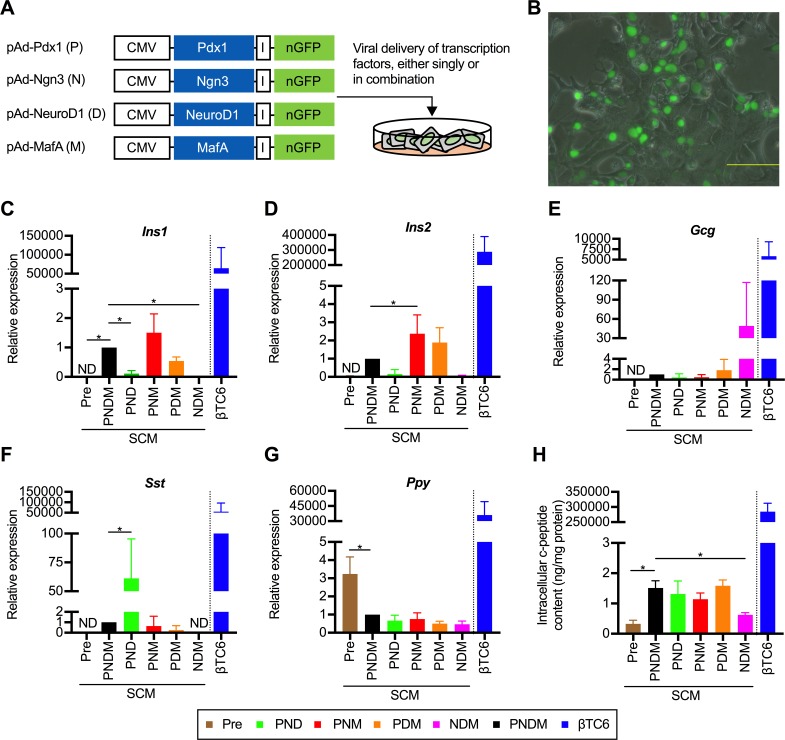
Fig 1. A triple combination of transcription factors (TFs) induces transdifferentiation into insulin-producing cells in vitro.
(A) Schematic diagram of the experimental strategy used to optimize the transgene. Adenoviruses encoding bicistronic TFs and nuclear localized GFP (nGFP) linked by an IRES element (I) were used. Each vector containing genes encoding Pdx1, Ngn3, NeuroD1, and MafA is abbreviated as P, N, D, and M, respectively. The combination of P, N, D, and M means the co-infection of each vector. (B) Appearance of nGFP-positive cells on the day after transfection. Scale bar, 200 μm. (C, D, E, F and G) Quantitative real-time polymerase chain reaction (qRT-PCR) analyses stratified according to the combination of TFs that were used were performed using primer sets for Ins1 (C), Ins2 (D), Gcg (E), Sst (F) and Ppy (G). “Pre” represents PBLHCs without transfection. The data are presented as fold-changes in the gene expressions relative to the values in the cells transfected with PNDM (n = 4). *P < 0.05 (one-way ANOVA followed by Dunnett’s test. The values in the cells transfected with PNDM were used as the control.); error bars, S.D. (H) Intracellular c-peptide contents in the transfected cells (n = 4). *P < 0.05 (one-way ANOVA followed by Dunnett’s test. The values in the cells transfected with PNDM were used as control.); error bars, S.D. The results obtained from βTC6, a mouse insulinoma cell line, were excluded from all the statistical analyses, because of the marked deviations from other values. CMV, cytomegaloviral promoter; Ins1, insulin I; Ins2; insulin II; Gcg, glucagon; Sst, somatostatin; Ppy, pancreatic polypeptide.