Fig. 2. Stn1 SIM mutations affect Stn1-Ten1 recruitment and enhance telomerase binding to telomeres.
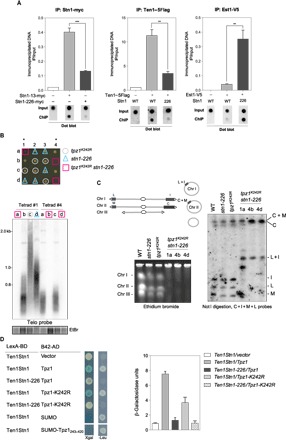
(A) Telomere association of Stn1, Ten1, and Est1 monitored with dot blot ChIP assays. ChIP experiments were performed using samples collected from 25°C exponentially growing WT stn1+ and stn1-226 cells carrying the indicated epitope-tagged proteins. Error bars indicate the SEM from multiple independent experiments (n = 3); P values are calculated from two-tailed t test (**P < 0.01 and ***P < 0.005). (B) Top: Growth of spores carrying the indicated genotypes. Bottom: Telomere length analysis of two tetrads by Southern blotting with a radiolabeled telomeric DNA probe. Genomic DNA was digested with Apa I restriction enzyme. (C) Left: PFGE stained by ethidium bromide (EtBr) of intact chromosomes from WT strain, parental tpz1K242R and stn1-226 single mutants, and tpz1K242R stn1-226 double mutants grown at 25°C. Chromosomes I, II, and III were stable in single mutants, whereas they did not enter into the gel in double mutants. Right: Not I–digested genomic DNA was subjected to PFGE, transferred to nitrocellulose membrane, and hybridized with probes of telomere proximal fragments C, I, M, and L. In tpz1K242R stn1-226 double mutants, these fragments were absent and have been replaced by DNA fragments C + M and L + I. This modification in the Not I–digested pattern is characteristic of chromosome circularization. (D) Ten1Stn1 interactions with Tpz1 analyzed by Y2H in different mutants. Cells were grown and spotted on selective medium (-TRP and -HIS) and tested for activation of Leu2 and/or LacZ reporters. Y2H interaction was quantified by measurement of β-galactosidase activity.
