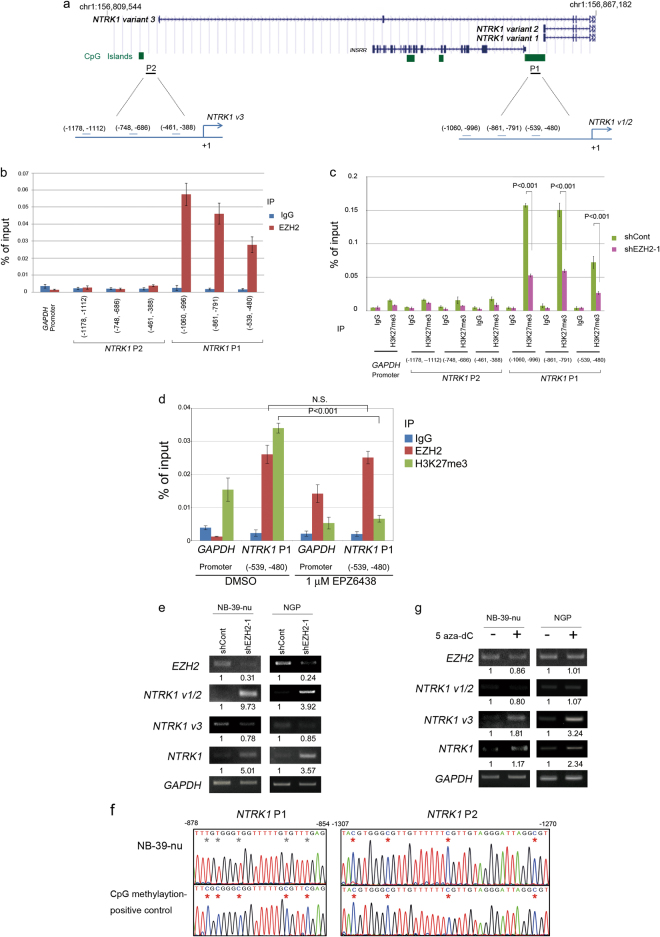
Fig. 6.
Complex epigenetic regulation of NTRK1 promoters by EZH2 and/or DNA methylation. a The positions of NTRK1 variants were displayed in the UCSC genome browser (Human GRCh38/hg38, https://genome.ucsc.edu). ChIP qPCR site positions in the two promoters of NTRK1, Promoter 1 (P1) and Promoter 2 (P2). b qChIP assay for EZH2 in NTRK1 P1 and P2 promoters in NB-39-nu cells. Immunoprecipitation was performed by an anti-EZH2 antibody and control rabbit IgG. Primer locations are indicated in the diagrams in a. Primer sequences are shown in Supplementary Table S4. Results are presented as fold enrichment and are representative of at least three independent experiments. c qChIP assay for H3K27me3 in NTRK1 P1 and P2 promoters in mock- and EZH2 knocked down NB-39-nu cells by the shEZH2-1 lentivirus. Immunoprecipitation was performed by an anti-H3K27me3 antibody and control rabbit IgG. d qChIP assay for H3K27me3 and EZH2 in the NTRK1 P1 core promoter region in mock- and EPZ6438-treated NB-39-nu cells. Immunoprecipitation was performed with an anti-EZH2 antibody, an anti-H3K27me3 antibody, and control rabbit IgG. e RT-PCR for NTRK1 variants 1/2 and 3 in EZH2 knockdown NB cells. f Bisulfite-sequence analysis of NTRK1 P1 and P2 promoters. Red and gray asterisks indicate methylated and unmethylated CpG sites, respectively. CpG methylase-treated DNA was used as a positive control. g RT-PCR for NTRK1 variants 1/2 and 3 in 5 aza-dC (5 μM)-treated NB cells