Abstract
A key prerequisite to achieve a deeper understanding of microbial communities and to engineer synthetic ones is to identify the individual metabolic interactions among key species and how these interactions are affected by different environmental factors. Deciphering the physiological basis of species–species and species–environment interactions in spatially organized environments requires reductionist approaches using ecologically and functionally relevant species. To this end, we focus here on a defined system to study the metabolic interactions in a spatial context among the plant-beneficial endophytic fungus Serendipita indica, and the soil-dwelling model bacterium Bacillus subtilis. Focusing on the growth dynamics of S. indica under defined conditions, we identified an auxotrophy in this organism for thiamine, which is a key co-factor for essential reactions in the central carbon metabolism. We found that S. indica growth is restored in thiamine-free media, when co-cultured with B. subtilis. The success of this auxotrophic interaction, however, was dependent on the spatial and temporal organization of the system; the beneficial impact of B. subtilis was only visible when its inoculation was separated from that of S. indica either in time or space. These findings describe a key auxotrophic interaction in the soil among organisms that are shown to be important for plant ecosystem functioning, and point to the potential importance of spatial and temporal organization for the success of auxotrophic interactions. These points can be particularly important for engineering of minimal functional synthetic communities as plant seed treatments and for vertical farming under defined conditions.
Introduction
Higher-level functions and population dynamics within microbial communities are underpinned by the interactions among the composing species within the community and their environment [1, 2]. Deciphering these interactions is a prerequisite to understand and manage complex natural communities [3] and to achieve community-level synthetic engineering [4–6]. To this end, increasing numbers of experimental studies and (meta)genomic surveys have shown that auxotrophic interactions, involving vitamins and amino acids, are widespread in many microbial natural communities [2, 7–10] and can also be engineered genetically to create synthetic communities [11–13]. Specific auxotrophic interactions among microbes are shown to influence ecosystem functioning; for example, infection outcomes within higher organisms [14], ecological population dynamics in the oceans [2], and the level of biodegradation of organic matter under anoxic conditions [15, 16].
It has been suggested that auxotrophies can result from reduced selective pressures for maintaining biosynthesis capabilities under stable metabolite availability due to abiotic or biotic sources [7, 8]. This proposition is supported by the observed independent evolution of vitamin and amino acid auxotrophies in different, unrelated taxa [17, 18], and points to a direct linkage between ecological dynamics and evolution of auxotrophies [15]. The possible fluctuations in metabolite availabilities in time and space would be expected to impact both the emergence of auxotrophies and the population dynamics of resulting auxotrophic species. For example, in the marine environment, where the observed auxotrophies relate mostly to the loss of biosynthesis capacity for vitamins and amino acids, population dynamics of auxotrophic species are believed to be directly linked to those of “provider” species [2, 7, 19]. The ecological influences of auxotrophic species on community structure and population dynamics can also be exerted by abiotic fluctuations or directly by the abundances and actions of higher organisms within the system.
These ecological influences on microbial population dynamics can increase significantly in spatially organized systems. Yet, the spatial context of microbial interactions is only beginning to be appreciated [10, 20–22]. Considering that each species can display multiple metabolic actions and that all of these can affect a common environment, it is not clear if auxotrophic interactions are always successfully established even if genetic/metabolic complementarity is present. For example, metabolic interactions among genetically engineered auxotrophic yeast are shown to require an initial autonomous growth period to establish [10]. Establishment of these metabolic interactions might also involve spatial factors including the formation of metabolic gradients and specific population organization might be important [15, 20–23]. These spatial factors can include oxygen and pH gradients, as both of these can significantly change upon growth of one species [24–27], and can directly influence subsequent or simultaneous metabolic interactions or growth of different species. Studies exploring the possible interplays between species–species and species–environment interactions have so far used synthetically engineered interactions [10, 20, 21] or enriched microbial communities [15]. Further analyses of such interplay in ecologically and biotechnologically relevant systems can thus help engineering of novel applications of microbial communities with inherent spatial organization, such as seen in agriculture and involving for example closed-ecosystem production, seed treatment, and microbe-based biofertilization [28–30].
Towards this goal, we focus here on identifying potential metabolic interactions among the plant-beneficial endophytic fungus Serendipita indica (previously called Piriformospora indica [31]) and the ubiquitous soil microbe Bacillus subtilis. Identifying a defined media for S. indica, we found it to be auxotrophic for the vitamin B1, thiamine. To study the potential auxotrophic interactions of S. indica, we then created a co-culture system using B. subtilis. We found that S. indica thiamine auxotrophy can be satisfied and its growth restored by B. subtilis. The success of this auxotrophic interaction, however, is strongly dependent on temporal and spatial organization in the system. These findings and the established synthetic co-culture can act as a basis to develop a more complete functional synthetic community, as advocated for biotechnological applications and for gaining insights into community function [4, 6, 32, 33]. With the inclusion of a plant, such a synthetic community can allow further insights into microbe–microbe, and microbe–plant interactions and development of new agricultural technologies such as in seed coating and vertical farming in controlled environments.
Results
Serendipita indica is auxotrophic for thiamine
Serendipita indica is an endophytic fungus that can colonize roots of a wide range of plants and can confer a range of beneficial effects, including enhanced plant growth, resistance to biotic and abiotic stresses [34–36], promotion of adventitious root formation in cuttings [37], and assisting phosphate assimilation [38]. Despite its broad host range, S. indica also has the ability to grow in the absence of host plants [39]. Exploiting this ability, we attempted to create a fully defined growth medium that was based on previous physiological studies on S. indica [39–43]. Using our defined medium, we tested the effect of the vitamins on growth by cultivating S. indica in a series of vitamin-free media each supplemented by a specific vitamin (Fig. 1). The results showed that S. indica is auxotrophic for thiamine; while none of the other individual vitamin additions supported growth, thiamine and full vitamin addition did. This finding was further confirmed by growing S. indica on plates supplemented with an additional agar block only containing a defined amount of thiamine. In this case, growth of S. indica resulted in expansion towards the thiamine agar block, suggesting that growth occurs on a thiamine gradient or is linked with an active chemotaxis towards the thiamine source (Supplementary Information; Figure S1). We also quantified the growth of S. indica with different concentrations of thiamine and found that hyphae growth and spore formation showed a positive, but saturating, dependency on thiamine levels (Supplementary Information; Figure S2). Without thiamine in the media, we still observed germination and very little hyphal growth (Supplementary Information; Figure S2), possibly supported by spore-stored thiamine. Besides these measurements, we also observed thiamine effects on S. indica growth using time-lapse microscopy (see discussion below and videos provided as Supplementary Files 1 and 2).
Fig. 1.
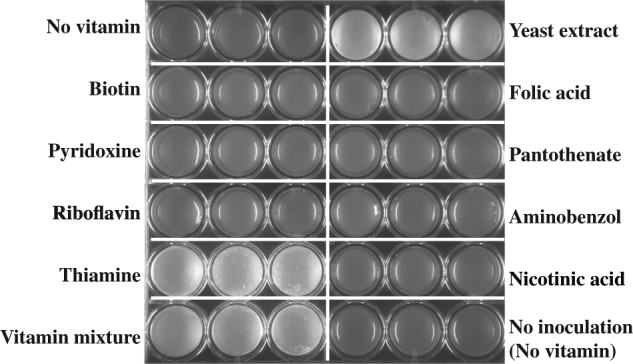
Serendipita indica growth under different conditions. Growth on agar plates with defined medium supplemented with different vitamins as shown on each row and column. Each treatment has three replicates presented in three adjacent wells. Serendipita indica grows in white colonies. Images were taken after 2 weeks of growth
Serendipita indica auxotrophy is reflected in its genomic enzyme content
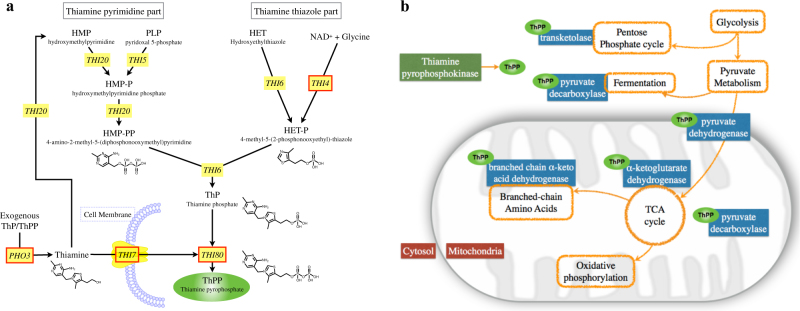
To support and better understand these physiological results, we analyzed the S. indica genome for the presence of genes associated with thiamine use, biosynthesis, and transport (Fig. 2a). This bioinformatics analysis revealed that S. indica lacks most of the genes of the thiamine biosynthesis pathway (Supporting Table S1 and Fig. 2a). In particular, we did not find any homologs of the genes THI5, THI6, and THI20. THI5 encodes the enzyme involved in the synthesis of the thiamine-precursor hydroxymethylpyrimidine (HMP), THI6 encodes the bifunctional enzyme acting as thiamine phosphate pyrophosphorylase and hydroxyethylthiazole kinase, and THI20 encodes an HMP kinase that displays both kinase and thiaminase II activity [44]. For the gene THI4, which encodes a bifunctional protein involved in thiamine thiazole synthase and mitochondrial DNA damage tolerance [45–47], we found a truncated homolog (11% length coverage against THI4 sequence in S. cerevisiae). Considering all other key biosynthesis genes (THI5, THI6, and THI20) are absent, it is possible that this truncated form relates to mitochondrial DNA damage tolerance [45–47]. We also found that S. indica contains a homolog of the THI7 (or alternative name THI10) that encodes a thiamine transporter, and a homolog of the PHO3 gene, whose product catalyzes dephosphorylation of thiamine phosphate to thiamine, thereby increasing its uptake [45]. The S. indica genome also contains THI80, which encodes a thiamine pyrophosphokinase catalyzing the conversion of thiamine into thiamine pyrophosphate (ThPP), the active form of thiamine as a key co-factor involved in central metabolic reactions [48]. In particular, ThPP is involved in pyruvate fermentation and conversion for entry into the citric acid cycle (TCA), α-ketoglutarate to succinyl-CoA conversion in the TCA cycle (Fig. 2b and S3), transketolase reactions in the pentose phosphate pathway, and biosynthesis reactions for leucine, isoleucine, and valine [49].
Fig. 2.
Thiamine-related genes and reactions. a Overview of the thiamine biosynthesis pathway in Saccharomyces cerevisiae based on Ref. [47], and as included in the KEGG metabolic pathways (Pathway: sce00730) [85]. Yellow boxes indicate genes encoding for the enzymes in the corresponding reactions. Red borders indicate genes for which there are S. cerevisiae homologs in S. indica (see Methods). b Simplified schematic of central metabolism in eukaryotic cells with cytosol and mitochondria compartments indicated. Orange enclosures show reactions of the central metabolism, while blue boxes show essential enzymes catalyzing these reactions and requiring ThPP as a co-factor (shown in green). Green oval indicates ThPP . Red boxes indicate the positions inside a cell
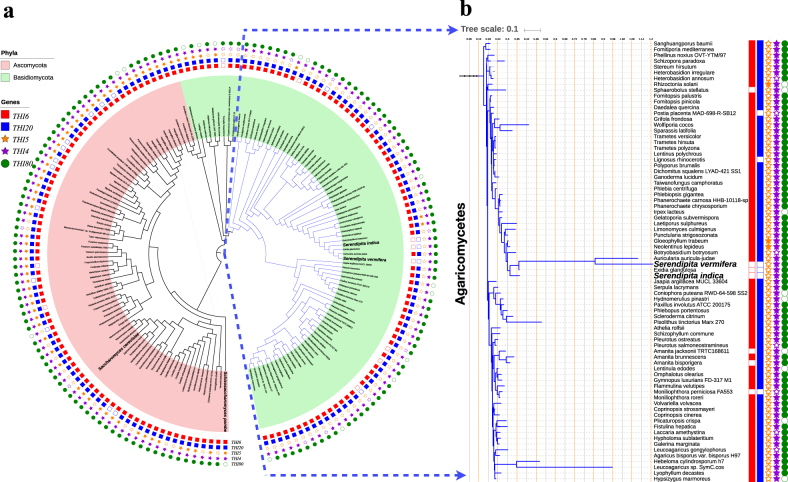
These results support the experimental observation of thiamine auxotrophy in S. indica. To see if such a loss of thiamine biosynthesis is widespread in other fungi, including evolutionarily closely and more distant relatives of S. indica and unrelated fungal species, we analyzed the absence and presence of all of the above key genes in available fungal genomes in the phyla of Basidiomycota (featuring S. cerevisiae) and Ascomycota (featuring S. indica). We mapped this information onto a phylogenetic tree that we created using the 18s ribosomal gene (see Methods). Summarized in Fig. 3, this analysis showed the same genetic pattern (including the truncated version of the THI4 homolog) for key thiamine biosynthesis genes in Serendipita vermifera, the closest relative of S. indica. In addition, we found that within the analyzed species from the Basidiomycota phylum, there are five species lacking either THI20 or THI6, and five species lacking both genes (Fig. 3), suggesting that they are unable to synthesize thiamine on their own (Fig. 2a). Among the analyzed species from the Ascomycota phylum, there are 14 species lacking either THI20 or THI6, while none of them lack both genes (Fig. 3). Taken together, these findings suggest that a complete or partial loss of thiamine biosynthesis genes can be observed in different fungi from different classes, rather than being confined to a specific class.
Fig. 3.
Summary of phylogenetic and bioinformatics analyses. a The phylogenetic tree of all 162 fungal species used (see Methods). Key species, S. cerevisiae, S. pombe, S. indica, and S. vermifera, are highlighted in bold. The tree contains 77 species from the phylum of Ascomycota (red background) and 85 species from the phylum of Basidiomycota (green background). Of the latter, 76 species are in the class of Agaricomycetes (blue branches). b Closer view of the phylogenetic relations among all 76 species in the class of Agaricomycetes. On both panels a and b, the presence/absence of each of the five thiamine biosynthesis genes in each species is indicated with different colors and symbols (as shown in the figure). Filled (empty) symbols indicate presence (absence) of a given species
Bacillus subtilis complements S. indica’s auxotrophy for thiamine and promotes its growth
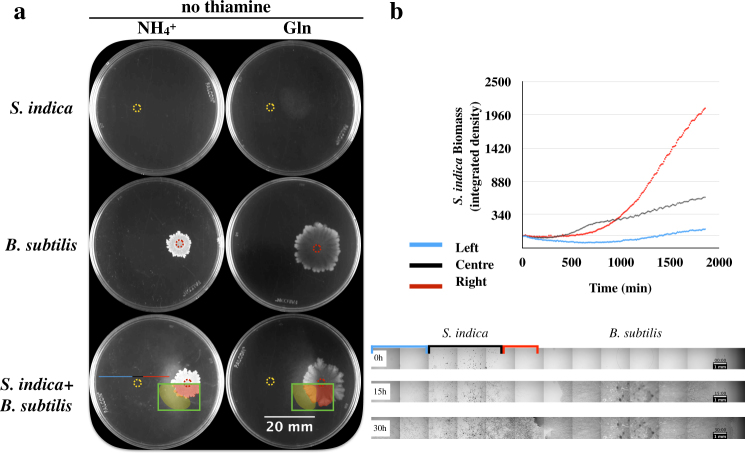
Given the crucial role of thiamine-derived co-factors in central metabolism (Fig. 2b), S. indica growth in nature apparently depends on environment-derived thiamine. Indeed, thiamine can be synthesized by various bacteria, fungi, and plants [50–52]. Among these, B. subtilis, a bacterium commonly found in the soil [53] is an established model organism [54]. Bacillus subtilis also requires thiamine for its growth and its thiamine biosynthesis pathways are well studied [50, 55]. Combined with the fact that B. subtilis is normally a plant-beneficial microbe [56], this motivated us to explore the possibility that the identified S. indica auxotrophy for thiamine could be satisfied upon co-culturing with B. subtilis. We created co-cultures of these two species on agar plates using our defined medium and two common nitrogen sources to evaluate possible auxotrophic interactions under these conditions. We found that B. subtilis could indeed stimulate S. indica growth under thiamine-free conditions and that S. indica growth followed a spatial pattern, with significant growth in the vicinity of the B. subtilis colony (Fig. 4a). A similar spatial growth was observed in experiments with S. indica and a supplied thiamine agar block (Supplementary Information; Figure S1).
Fig. 4.
Serendipita indica and B. subtilis interactions on agar plates. a Rows from top to bottom show growth of monocultures of S. indica, B. subtilis, and their co-culture, respectively, under the absence of thiamine. The yellow dotted circle on the images indicates the S. indica inoculation point. Red dotted circle indicates B. subtilis inoculation point. The left and right columns show growth on plates after 2 weeks, using ammonium and glutamine as nitrogen sources, respectively. Green square highlights the colonies with pseudo-color, where the area covered by S. indica hyphae is shown in yellow, and the B. subtilis colony is shown in red. The areas were manually drawn. When both organisms were cultured together (bottom row), B. subtilis and S. indica were inoculated on the right and left of the plate, respectively. Plates shown are representative of at least three replicates for each condition. We performed two biological replicates of this experiment, with qualitatively similar results. b, Top: S. indica biomass obtained from the defined horizontal sections of the plate of S. indica and B. subtilis co-culture on thiamine-free medium (the bottom left plate shown in the third row of part a). Biomass was calculated by integrated density of each horizontal sections based on image analysis. The blue, black, and red lines correspond to the horizontal sections as shown on the plate and in the time-lapse image series of S. indica and B. subtilis growth on agar plates as below. Bottom: The microscopy images are from the middle cross-section of the plate through the inoculation point of S. indica and B. subtilis, at 0, 15, and 30 h after inoculation. The red, black, and blue highlighted sections correspond to the S. indica colony side closer to B. subtilis, the middle of the colony, and the colony side far from B. subtilis, respectively
We used time-lapse microscopy to quantify this spatial growth pattern of S. indica and found that growth (as approximated by image density) happened faster at the side closer to the B. subtilis colony compared to the far side of the plate (see Fig. 4b and videos provided as Supplementary Files 3 and 4). This could be explained by the presence of an increasing thiamine gradient towards the B. subtilis colony that facilitates S. indica hyphal growth. While these findings strongly suggest a B. subtilis-linked thiamine provision, which then promotes S. indica growth, our attempts to quantify thiamine from agar plate co-cultures has failed, presumably due to a combination of thiamine consumption and sensitivity limitations of available thiamine assays (50 μg/l) [57]. We were, however, able to detect thiamine from concentrated B. subtilis cultures, and found the concentration in liquid culture supernatants to be approximately 7.56 μg/l.
While the above findings strongly suggest that the growth enabling of S. indica by B. subtilis is due to thiamine supply, it is theoretically possible that B. subtilis provides metabolites other than thiamine, which allow bypassing of central reactions requiring ThPP as a co-factor. In other words, provision of metabolites that are downstream of pyruvate in the TCA cycle (Fig. 2b). To examine this possibility, we analyzed growth of S. indica in the absence of thiamine but supplemented with organic and amino acids that link to the central carbon metabolism. We found that none of the 17 amino acids or 8 organic acids tested or their combinations allowed for S. indica growth in the absence of thiamine (Supplementary Information; Figure S4). This finding further confirmed that B. subtilis facilitated growth of S. indica in thiamine-free medium is linked directly to thiamine.
Metabolic profiling shows additional metabolic interactions between S. indica and B. subtilis
To analyze the basis of metabolic interactions between the two organisms and to collect more evidence for thiamine-based auxotrophy, we grew each organism in liquid culture on its own and then cross-cultured the other organism on the supernatant of the first one. As with agar plates, we found that in the absence of thiamine, the S. indica growth was limited to spore germination (Fig. 5). When supplemented with B. subtilis supernatant, however, S. indica showed significantly increased growth in liquid culture (Fig. 5). There was also a growth enhancement of S. indica by the B. subtilis supernatant when cultured in the presence of thiamine.
Fig. 5.
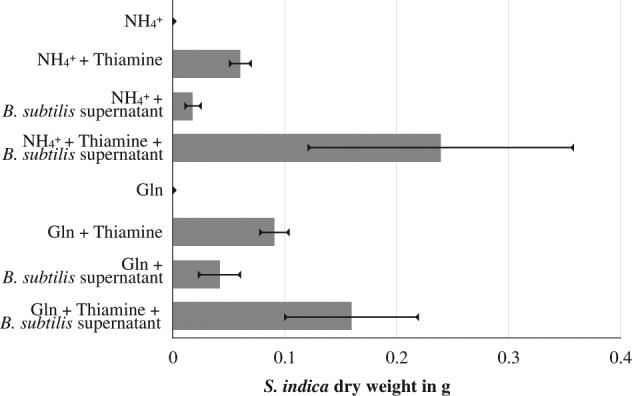
Serendipita indica growth under different media compositions. Growth using either ammonium or glutamine and supplemented with thiamine or B. subtilis supernatant, as indicated on the y-axis. The x-axis shows S. indica growth approximated by total dry weight after 1 week of growth. Three technical replications were used for generating the weight data. Data shown here is from one of the two biological replications, with others giving quantitatively similar results
To further elucidate these physiological observations, we repeated these experiments and undertook an ion chromatography (IC)-based targeted metabolite quantification (see Methods). In particular, we were able to quantify the key nitrogen compounds (ammonium, glutamine, and glutamic acid), and organic and amino acids relating to the TCA cycle (alanine, glycine, lactate, acetate, pyruvate, and formate) (see Fig. 2 and S3) in the supernatant of each organism before and after cross-cultivation. We found that the supernatant from B. subtilis monoculture contained significantly higher amounts of acetate and some formate and pyruvate, and that the extracellular levels for these compounds did not change in the presence or absence of thiamine in the media (Supplementary Information; Figure S5). When S. indica was grown in the B. subtilis supernatant and in the absence of thiamine, the fungus consumed both acetate and formate and produced pyruvate (Supplementary Information; Figure S5). In the presence of both thiamine and the B. subtilis supernatant, the consumption of acetate and formate was also observed, but there was also production of lactate in addition to pyruvate (Supplementary Information; Figure S5). There were also few peaks in the IC chromatograms that are altered during the cross-supernatant experiment and that could not be identified (Supplementary Information; Figure S6). In the case of the key nitrogen compounds and amino acids, we found that in glutamine media, B. subtilis supernatant contained glutamic acid and glutamine, which were subsequently consumed by S. indica (Supplementary Information; Figure S7). In ammonium-based media, we found significant amounts of ammonia unconsumed in the B. subtilis supernatant, which is then subsequently consumed by S. indica. This ammonia consumption by S. indica led to the accumulation of glutamic acid, glutamine, alanine, and small amounts of glycine (Supplementary Information; Figure S7).
These findings, in particular acetate and formate cross-feeding from B. subtilis to S. indica, explain the positive impact of B. subtilis supernatant on growth irrespective of thiamine availability. They also provide further support that the B. subtilis-associated growth of S. indica relates to thiamine provision rather than organic acids, since acetate and formate alone did not enable S. indica growth in thiamine-free media (Supplementary Information; Figure S4).
The successful co-existence of S. indica and B. subtilis depends on spatiotemporal organization in the system
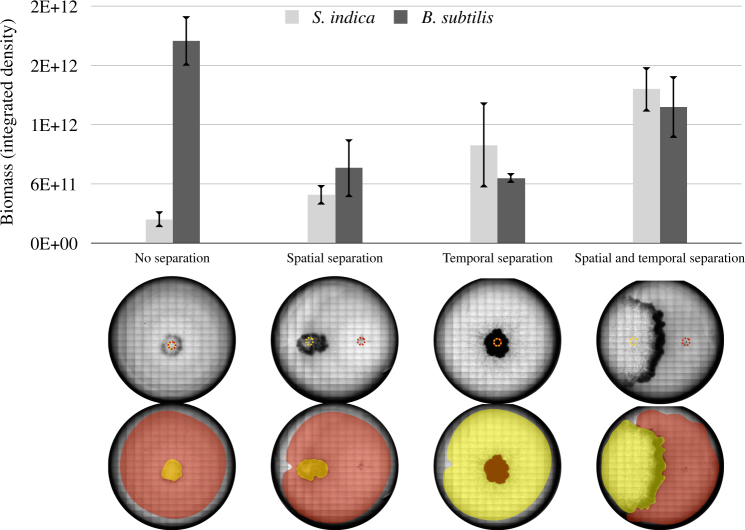
The above findings show that B. subtilis can support the growth of S. indica in thiamine-free medium either through its supernatant or when co-cultured at a distance on an agar plate. Both experimental setups were geared towards identifying possible interactions among the two species through utilization of the excretions of one species by the other, but did not necessarily consider the spatiotemporal factors on such interactions. Thus, the remaining question was whether both species could still co-exist and establish a successful interaction under different conditions regarding the spatial proximity or size of initial inoculation, or the actual growth phase that the different species are in at the time of introduction onto the agar. While addressing these questions is experimentally challenging, we attempted here to analyze the impact of spatial-temporal factors on the outcome of the S. indica–B. subtilis auxotrophic interaction by changing inoculation time and location on agar plates. In particular, we separated the inoculation of S. indica spores from B. subtilis inoculation either in time (by inoculating spores 3 days before B. subtilis colony inoculation) or space (by inoculating S. indica and B. subtilis at certain distance to each other) (see Methods). Alternatively, we inoculated S. indica spores after mixing with B. subtilis. These experiments mimic a scenario commonly found in agrotechnology practices when using pre-mixed cultures or spores of different microbes as soil biofertilizers or plant seed pre-treatments [58]. We found that with temporal or spatial separation, both species could successfully grow in the absence of thiamine, indicating a positive auxotrophic interaction (Fig. 6). In contrast, direct co-inoculation of B. subtilis with S. indica significantly hampered co-existence of the two species, particularly reducing S. indica growth (Fig. 6).
Fig. 6.
Biomass of B. subtilis and S. indica under different spatiotemporal culturing cases. “No separation” refers to B. subtilis culture and S. indica spores being pre-mixed at 1:1 volume ratio, and then inoculated as a single solution. “Spatial separation” refers to approximately 1.5 cm separation of S. indica (left) and B. subtilis (right) inoculation points. “Temporal separation” refers to inoculation of S. indica 3 days prior to B. subtilis inoculation. The yellow dotted circle on the images indicates the S. indica inoculation point. Red dotted circle indicates B. subtilis inoculation point. Below the graph are shown the plate images. The pseudo-coloring indicates S. indica hyphal area (in yellow) and B. subtilis colony (in red). The area of each colonies was manually drawn. Growth of the different species was approximated by tracing their respective colonies on the plate and measuring the image intensity from the engulfing areas 2 weeks after S. indica inoculation. Measurements are from three replicate agar plates, with a representative plate image shown at the bottom. These images show microscopic scans of each plate at 2 weeks of growth. For the “no separation” case, there was no observable S. indica colony expansion after 1 week. We performed four biological repetitions of this experiment, with qualitatively similar results
One possible explanation for this observed strong effect of co-inoculation is that alterations of the micro-environment by one species causes inhibitory effects on the other. Indeed, previous studies have shown that microbial growth on agar plates can create oxygen depletion within a colony [24, 25, 59]. This explanation could be particularly relevant in our experiments, as germination and initial growth of some soil fungi are shown to require substantial oxygen [60]. To gain insight on oxygen levels in our experimental system, we have repeated the experiments shown in Fig. 6, and measured oxygen levels on the agar plate (see Methods). We could confirm, as observed before [59], that oxygen is completely depleted on the B. subtilis colonies, already within few hundred microns underneath the surface of the plate/colony (Figure S8). In contrast, oxygen levels on the inoculation and growth zone of S. indica, as well as elsewhere on the agar plate remained high to significant depths from agar/colony surface (Supplementary Information; Figure S8). These results support the hypothesis that oxygen depletion by B. subtilis might inhibit germination of S. indica spores in “pre-mixed” inoculations.
Discussion and conclusions
Here, we report an auxotrophy for thiamine in the endophytic fungus S. indica and how this thiamine requirement can be satisfied by B. subtilis. We show that S. indica growth in thiamine-free medium cannot be supported by any other vitamin or relevant organic and amino acid, but can be restored when co-cultured with B. subtilis or with its liquid culture supernatant. While these co-culturing experiments identified additional metabolic interactions between these two species, they support a primary and successful auxotrophic interaction between the two organisms.
The presented experimental findings on thiamine auxotrophy are supported by bioinformatics analyses, showing that S. indica and its close relative S. vermifera lack the key thiamine biosynthetic genes THI5, THI6, and THI20, and only contain a truncated version of the THI4 gene. Instead, both organisms possess gene homologs encoding for proteins involved in the efficient transport of thiamine and its conversion into an active form (THI7, PHO3, and THI80, respectively). A phylogenetic analysis shows that these genetic patterns are not unique to these two species, but also not widespread, indicating that loss of thiamine biosynthesis can be related to their ecology and life style (both species are general plant colonizers [31]. Within an environment where thiamine is readily available, it is indeed possible that thiamine biosynthesis is lost due to enzymatic costs. In particular, the protein product of THI4 is shown to undergo only a single turnover before requiring re-synthesis, a so-called “suicide enzyme” [61]. Due to this potential high cost, loss of THI4-related thiamine biosynthesis might be adaptive under circumstances where thiamine can be acquired from the environment [48]. In both S. indica and its close relative S. vermifera genome there is only a truncated THI4 gene (11% coverage against THI4 sequence in S. cerevisiae), and it is possible that this gene relates only to the known additional function in mitochondrial DNA repair [46, 61] rather than thiamine biosynthesis (as the other thiamine biosynthesis THI5, THI6, and THI20 are absent).
Interestingly, we found that the auxotrophic interaction between S. indica and B. subtilis can only be achieved under conditions where the inoculation (and germination) of the two species is separated in time or space. This finding adds to an array of recent studies demonstrating the importance of spatiotemporal organization for the emergence and maintenances of metabolic interactions in microbial and other systems [15, 20–23]. In general, such effects can emerge from a multitude of factors including competition for local resources, spatial factors such as mixing and physical contact, or changes in ecological conditions caused by species' activities. While a detailed elimination of each possible factor might be difficult in specific cases, our findings suggest that in the case of S. indica and B. subtilis, oxygen depletion by a growing B. subtilis colony can cause inhibition of S. indica seed germination, and result in the failure of any auxotrophic interaction from establishing. Observations on oxygen depletion in growing bacterial colonies has been made before [24, 25, 59], as well as the idea that changing micro-environment conditions can influence the establishment and maintenance of microbial interactions [10, 15, 23, 62, 63].
Emerging studies have revealed the extent and importance of metabolic interactions within co-cultures and larger microbial communities using metabolomics approaches [9, 10, 63]. In the presented study, we focused on characterizing the co-culture dynamics given the primary auxotrophic interaction via thiamine. We have also used a targeted metabolomics approach to identify additional metabolic interactions, involving key organic and amino acids related to primary metabolism (in particular the TCA cycle). These additional interactions hint to the role of overflow fluxes from central metabolism in creating new or additional metabolic interactions that can operate on top of auxotrophic interactions. It is also possible that these different types of interactions can facilitate each other’s emergence and stabilization. For example, in the case of thiamine auxotrophy, limitations in thiamine levels can decrease the rates of pyruvate and α-ketoglutarate processing through the TCA cycle, leading to diversion into amino acids such as glutamic acid, glutamine, alanine, and glycine (see Supplementary Information; Figure S3), which can then create new metabolic interactions. Thus, overflow metabolism and its relation to the availability of electron acceptors or co-factors could provide a conceptual framework to understand and study microbial interactions [64, 65]. This view is also supported by recent studies indicating the role of both carbon and nitrogen overflows in the establishment of metabolic interactions [23, 63, 66].
The presented findings have implications both for the study of S. indica, as an important plant-supporting soil fungus [41], and for the engineering and application of minimal synthetic communities that aim to establish plant-supporting soil communities. In the former direction, future metabolic and physiological studies of S. indica will be enabled by the defined media conditions and identified thiamine auxotrophy in this study. A key suggestion from a biotechnological perspective, for example, is to consider thiamine as an important factor in the commercial mass production of S. indica [67]. In the latter direction, the finding that the success of auxotrophic interactions relates to spatiotemporal effects suggests that consideration should be given to inoculation timing when designing or applying biofertilizers or bio-control agents to the soil. Indeed, microbial interactions and synergisms are suggested to be crucial for soil fertility, bioproductivity, and ecosystem functioning [68–70]. Plants significantly benefit from symbioses with soil microbes, with benefits ranging from nutrient supply, growth promotion to elevating plant stress resistance [56, 71–74]. At the same time, soil microbes can interact among themselves or alter each other’s interactions with the plants [75–78]. The biochemical basis of these potential multi-level interactions in the soil has remained mostly elusive to date, with few documented cases of amino acid auxotrophies in specific soil bacteria and vitamin provision from plants relating to their root colonization [79–81]. The presented synthetic community of S. indica and B. subtilis shows that metabolic auxotrophy can directly underpin microbial interactions and growth, and that the success of interactions can be determined by spatiotemporal organization in the system. This synthetic system allows controlled investigations (and potential optimization) of fungal–bacteria interactions and can be further extended with additional microbes and a plant. The resulting minimalist synthetic ecosystem can provide a platform to analyze and control cross-kingdom relationships between plants and their growth-promoting fungi and bacteria [90], and enable new applications for vertical farming and crop production in the future.
Materials and methods
Serendipita indica cultures, growth media, and conditions
The defined basic medium for testing S. indica growth with ammonium as nitrogen source contained per liter; 15 g agar, 20 g glucose, 1.32 g (NH4)2SO4, 0.89 g Na2HPO4·2H2O, 0.68 g KH2PO4, 35 mg Na2MoO7·2H2O, 5.2 mg MgSO4, 2.5 mg FeSO4·7H2O, 0.74 mg CaCl2·2H2O, 0.0043 mg ZnSO4·7H2O, and 0.004 mg CuSO4·5H2O. Growth experiments for testing effects of different vitamins were performed in 24-well plates (Ref: 353226, Falcon), where each well contained 2 ml of the basic medium, supplemented with either 200 μg/l of single vitamin solution, 1 g/l yeast extract, 200 μg/l mixture of all eight vitamins tested, or equivalent amount of deionized water. Each well was then inoculated with 1 μl of S. indica spore suspension (approximately 500,000 spores/ml, counted with Neubauer counting chamber), where spores were harvested from 6 to 8 weeks old S. indica agar plates. In the “non-inoculated” control treatment, 1 μl of deionized water was used instead. Each treatment condition was prepared in three technical replicates. The 24-well plates were then sealed with parafilm and placed in a 30°C static incubator for 2 weeks. Images were taken with a gel doc system (Syngene) at the end of this period.
Experiments on agar plates, growth media, and conditions
The defined (basic) medium for testing S. indica growth on agar plates is given as above. When the chosen nitrogen source was ammonium, 1.32 g/l (NH4)2SO4 was added to this basic recipe. When the chosen nitrogen source was glutamine, 1.46 g/l glutamine was added to this basic recipe. For experiments with thiamine, 150 μg/l thiamine was added to the basic media after autoclaving.
Experiments were carried out on 60 mm dishes, filled with 6 ml of agar medium given above. A 500,000 spores/ml S. indica spore suspension, with spores harvested from 6 to 8 weeks old S. indica agar plates, was inoculated with 2 μl (on the left side of the plates). At approximately 2 cm distance to the right of the inoculum, either a “mock” solution, a thiamine block or a B. subtilis inoculum were placed. The “mock” solution was 2 μl of sterile water. The thiamine blocks were made by pouring 6 ml of 1.5% agar solution containing150 μg/l thiamine into a 60 mm plate, and then punching a block out using a sterile pipette tip with a diameter of 4.7 mm. Therefore, each agar block used contained approximately 5.6 ng thiamine. The B. subtilis inoculum was a 2 μl sample harvested from a culture, grown in 5 ml liquid Lysogeny broth (LB) [83] to an OD600 ≈ 0.5 measured by spectrophotometer (Spectronic 200, Thermo Fisher Scientific), then washed and re-suspend to OD600 ≈ 0.5 with 10 mM MgCl2. The plates were incubated in 30°C for 2 weeks. Images were taken with gel doc system (Syngene) at the end of this period.
Time-lapse microscopy and image analysis
Time-lapse microscopy was performed on agar medium cultures that were prepared using the same basic medium described above. A 6-well tissue culture plate (Ref: 353046, Falcon) was used and 1 μl of S. indica, B. subtilis or mock solution prepared as described above were inoculated on each well accordingly to experiment design. An Olympus IX83 microscope, UPlanFLN ×4 objective and cellSens software were used for recording the growth. Okolab stage top incubator (H301-T-UNIT-BL-Plus system, and H301-EC chamber) were used for incubation, with a temperature sensor and lens heater set to 30°C and stabilized for at least 2 h prior to the experiment. Different fields of view were chosen at interior and periphery of each colony and images from those fields were recorded using the automated microscope stage and Olympus cellSens software. Images were taken in 1 h intervals and put together as image series. ImageJ (Fiji) [84] was used for measuring the mean intensity on each field of view over time, normalized against the intensity value of the first frame from each view point (as shown in Fig. 4).
Spatial and temporal separation experiments
Serendipita indica (500,000 spores/ml, determined by counting with Neubauer counting chamber) and B. subtilis (OD600 ≈ 0.5, determined by spectrophotometer (Spectronic 200, Thermo Fisher Scientific)) were cultivated on thiamine-free synthetic medium containing ammonium as sole nitrogen source, and using six-well tissue culture plates (Ref: 353046, Falcon). On each plate, 1 μl of S. indica and 1 μl of B. subtilis were inoculated on 5 of the wells; one well was intentionally left non-inoculated as a blank. In the “no separation” case, S. indica and B. subtilis were pre-mixed at 1:1 volume ration, and inoculated on the center of each well. For “spatial separation” case, S. indica was inoculated 7.5 mm left to the center of a well and B. subtilis 7.5 mm right to the center, leaving 15 mm distance in between. For “temporal separation” case, S. indica was inoculated on each well, the plates were then incubated for 3 days and B. subtilis was inoculated after this time. The 3-day time separation was primarily based on the germination time of S. indica spores, which can take 2–3 days in our synthetic medium. As shown in the video included as Supplementary File S1, S. indica is able to germinate and grow a low amount of hyphae, potentially with available thiamine stored in spores. All the plates were incubated in 30°C for 2 weeks (starting from the time of S. indica inoculation). Images were taken by scanning each well under a microscope (Olympus IX83) using the same exposure time under bright field. ImageJ was used for measuring the biomass by integration of the total colony density. An image of each colony was manually outlined using the selection tool. The selected area was compared with the same location on a blank well from the same plate. The area and relative intensity were recorded (using “measure” function) and used for calculating the colony growth.
Oxygen measurements
Oxygen measurements were done in a replicate experiment with the “spatial separation” and “no separation” treatments described above and using with thiamine-free agar medium containing glutamine as sole nitrogen source. UniSense micro-electrode (OX-NP with Unisense microsensor multimeter and Unisense Sensor Trace Suite software) was used for daily oxygen measurements. Three identical plates of the same treatment and initiated on the same day was used for each measurement (i.e., three plates per day per treatment) and discarded afterwards. On each day, the micro-electrode was calibrated using aerated distilled water (air bubbled through for 3–5 min) and an anaerobic solution (prepared as per the manufacturer's instructions; 0.1 M ascorbic acid in 0.2 M NaOH, mixed briefly to dissolve and left unagitated for calibration 5 min prior to first measurement). For each measurement, the electrode was first placed on the colony or agar surface. Once a measurement at the surface was done, the electrode was lowered into the agar or colony by means of a manual micro-manipulator (Scientifica), and at 400 μm intervals in the z-dimension of the manipulator. Three depths (0.4, 0.8, and 1.2 mm from the surface) were used for oxygen measurement. Measurements were taken on B. subtilis, S. indica, and pre-mixed co-culture inoculation points, as well as on an off-location without any bacteria or fungi growth.
Supernatant cross-feeding experiments and metabolite analysis
Axenic cultures of S. indica and B. subtilis were cultivated in 50 ml basic medium described above, with or without thiamine. For S. indica an inoculum of 50 μl of a 500,000 spores/ml spore suspension, harvested from 6 to 8 weeks old S. indica agar plates, was used. For B. subtilis, an inoculum of 50 μl, sampled from a culture grown in LB to an OD600 ≈ 0.5 determined by spectrophotometer (Spectronic 200, Thermo Fisher Scientific), and washed with 10 mM MgCl2, was used. After 1 week incubation in 30°C and at 150 rpm, S. indica cells were harvested by centrifugation at 18,000 × g for 20 min. The supernatant was collected and filtered through a 0.2 μm polyethersulfone (PES) filter (Ref: WHA67802502, Whatman), while biomass was washed with 40 ml MilliQ water, dried using a centrifugal evaporator (EZ-2 Elite, Genevac), and then weighted. The growth of B. subtilis liquid cultures was monitored daily by taking 1 ml samples and measuring OD600 by spectrophotometer (Spectronic 200, Thermo Fisher Scientific). At the end of 1 week, the remaining liquid culture was centrifuged at 18,000 × g for 10 min. The supernatant was collected and filtered through a 0.2 μm PES filter, while biomass was discarded.
Both supernatants were mixed with fresh basic medium in a 1:1 ratio to set up new axenic cultures of S. indica and B. subtilis. One milliliter of liquid samples were collected from these cultures after 1 week by filtering through a 0.2 μm nylon membrane (Ref: WHA7402004, Whatman). Samples were transferred into polypropylene vials (Ref: 079812, Thermo Fisher Scientific) for IC, which was performed using Dionex ICS-500+ and column Dionex IonPac AS11-HC-4 μm (2 × 250 mm).
The same samples were sent for commercial amino acid analysis (performed by Genaxxon, Ulm, Germany). A polymeric cation exchanger was used to separate amino acids by high-performance liquid chromatography chromatography (particle size: 5 μm; column dimensions: 125 × 4.6 mm ID) (Amino Acid Analyzer LC3000). Separated amino acids were detected by post-column Ninhydrin derivatization at 125 °C and photometric measurement at 570 nm.
Sequence analysis and BLAST search of key thiamine biosynthesis genes
Sequences of key thiamine biosynthesis enzymes (Table S1) from Saccharomyces cerevisiae S288c were compared with available S. indica genome homologs using Position-Specific Initiated BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), to identify the putative functions of the corresponding genes. An evalue of 1e−6 was chosen as cut-off to identify homologous sequences [85]. The results of the analysis are shown in Table S1.
Phylogenetic and bioinformatics analysis
In order to understand the loss of thiamine biosynthesis function in S. indica, we performed a phylogenetic analysis of 162 fungi species including S. indica and its close relative S. vermifera, and focusing on thiamine biosynthesis genes. This focus meant that we can only use species with fully sequenced and well-annotated genomes. To collate fungi species fulfilling these criteria, we first used the KEGG database [86] in which there are 102 species with annotated genomes that are mapped to metabolic pathways. This dataset, however, contains only 16 species in the Agaricomycetes class, in which the species S. indica and S. vermifera belong. To get more close relatives of S. indica, we used the NCBI Taxonomy [87] and SILVA rRNA [88] databases and collated 60 species that are from the Agaricomycetes class and that have full genome assembly information. For the resulting 162 species, we downloaded SSU-rRNA sequences from SILVA rRNA database [88]. Where multiple sequences were present for a given species, we used the longest sequence with highest quality. We then aligned the resulting 162 rRNA sequences using the MUSCLE tool [89], and built a maximum-likelihood phylogenetic tree using PhyML [90]. Onto the resulting phylogenetic tree, we mapped the presence and absence information of the thiamine biosynthesis genes THI6, THI20, THI5, THI4, and THI80 (see Fig. 3). This information was already available for the 102 species that are contained in the KEGG database. To obtain it for the additional 60 species from the Agaricomycetes class (and including the species S. indica and S. vermifera), we ran a BLAST analysis, in which we queried each of these species genomes against the THI6, THI20, THI5, THI4, and THI80 gene sequences from S. cerevisiae. A gene was considered present in a given genome if there was a match of homologous sequences with an e-value smaller than 1e−6 [85].
Thiamine measurements on B. subtilis liquid cultures
Axenic cultures of B. subtilis were cultivated in 50 ml synthetic medium containing glutamine as a sole nitrogen source without thiamine. Bacillus subtilis inoculum of 50 μl, sampled from a culture grown in LB to OD600 ≈ 0.5 determined by spectrophotometer (Spectronic 200, Thermo Fisher Scientific), and washed with 10 mM MgCl2 was used. After 1 week of incubation in 30°C and at 150 rpm, B. subtilis cultures were harvested by centrifugation at 18,000 × g for 10 min. The supernatant was collected and filtered through a 0.2 nm PES membrane filter (Ref: WHA67802502, Whatman). The supernatant of 250 μl from each culture was transferred to a clean 1.5 ml Eppendorf tube, followed by sequentially adding 10 μl 1% K3[Fe(CN)6], 150 μl 15% NaOH solution, and 150 μl isobutanol. The tubes were shaken vigorously for 1 min, followed by 2 min of centrifugation at 13,000 × g. The upper isobutanol layer of each tube was transferred to a new 1.5 ml Eppendorf tube, containing approximately 0.2 g Na2SO4. The tubes were mixed well and centrifuged for 1 min at 13,000 × g for solids to settle. One hundred microliters of supernatant from each tube were transferred to 96-well plates (Ref: 3916, black flat bottom, Corning) and the fluorescence was measured using a plate reader (CLARIOstar, BMG Labtech) at 365 nm excitation and 450 emission. The concentration of thiamine was determined with a series of known concentration standard thiamine solutions under the same treatment.
Serendipita indica growth under different thiamine concentrations
Synthetic medium containing ammonium as sole nitrogen source was used for testing S. indica growth in different thiamine concentrations. Media containing final thiamine concentrations of 0, 1.5, 15, 150, and 1500 μg/l were prepared, and distributed in a 6-well tissue culture plate (Ref: 353046, Falcon). Each 6-well plate contained one concentration condition, with 3 ml medium in each well. On each plate, 1 μl of S. indica (500,000 spores/ml) were inoculated on the center of five wells, while one well was intentionally left non-inoculated as a blank. Plates were incubated at 30°C for 2 weeks. Afterwards, lids were removed and OD600 and fluorescence at 365 nm for excitation and 450 nm for emission were measured using a plate reader (CLARIOstar, BMG Labtech) and the plate scan function to get an overall reading of each well. Images of plates were taken using a gel doc system (Syngene).
Electronic supplementary material
Supplementary Information (containing Supplementary Figures and Tables)(PDF 6688 kb)
Acknowledgements
We would like to thank Sarah Bennett at the WISB research facilities for her help in IC analyses, and Chrysi Sergaki for her help with S. indica spore preparations.
Funding
This work is funded by research grants from Biotechnological and Biological Research Council (BBSRC) of the United Kingdom, with grant IDs: BB/K003240/2 (to OSS) and BB/M017982/1 (to the Warwick Integrative Synthetic Biology Center, WISB).
Author contributions
MA, PS, and OSS designed experiments and initiated research project. XJ performed experiments and analyzed data. CZ, SF, and RE provided resources and technical assistance. All authors contributed to the writing of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0095-z) contains supplementary material, which is available to authorized users.
Contributor Information
Patrick Schäfer, Phone: +44(0)2476575244, Email: P.Schafer@warwick.ac.uk.
Orkun S Soyer, Phone: +44(0)2476572968, Email: O.Soyer@warwick.ac.uk.
References
- 1.Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth’s biogeochemical cycles. Science. 2008;320:1034–9. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 2.Sañudo-Wilhelmy SA, Gómez-Consarnau L, Suffridge C, Webb EA. The role of B vitamins in marine biogeochemistry. Ann Rev Mar Sci. 2014;6:339–67. doi: 10.1146/annurev-marine-120710-100912. [DOI] [PubMed] [Google Scholar]
- 3.Abreu NA, Taga ME. Decoding molecular interactions in microbial communities. Shen A (ed) FEMS Microbiol Rev. 2016;40:648–63. doi: 10.1093/femsre/fuw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Großkopf T, Soyer OS. Synthetic microbial communities. Curr Opin Microbiol. 2014;18:72–77. doi: 10.1016/j.mib.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hays SG, Patrick WG, Ziesack M, Oxman N, Silver PA. Better together: engineering and application of microbial symbioses. Curr Opin Biotechnol. 2015;36:40–49. doi: 10.1016/j.copbio.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Lindemann SR, Bernstein HC, Song HS, Fredrickson JK, Fields MW, Shou W, et al. Engineering microbial consortia for controllable outputs. ISME J. 2016;10:2077–84. doi: 10.1038/ismej.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helliwell KE, Wheeler GL, Smith AG. Widespread decay of vitamin-related pathways: coincidence or consequence? Trends Genet. 2013;29:469–78. doi: 10.1016/j.tig.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Morris JJ, Lenski RE, Zinser ER. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. MBio. 2012;3:e00036-12. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romine MF, Rodionov DA, Maezato Y, Osterman AL, Nelson WC. Underlying mechanisms for syntrophic metabolism of essential enzyme cofactors in microbial communities. ISME J. 2017;11:1434–46. doi: 10.1038/ismej.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell K, Vowinckel J, Mülleder M, Malmsheimer S, Lawrence N, Calvani E, et al. Self-establishing communities enable cooperative metabolite exchange in a eukaryote. Elife. 2015;4:115. doi: 10.7554/eLife.09943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mee MT, Collins JJ, Church GM, Wang HH. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci USA. 2014;111:E2149–56. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pande S, Merker H, Bohl K, Reichelt M, Schuster S, de Figueiredo LF, et al. Fitness and stability of obligate cross-feeding interactions that emerge upon gene loss in bacteria. ISME J. 2014;8:953–62. doi: 10.1038/ismej.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell K, Vowinckel J, Ralser M. Cell-to-cell heterogeneity emerges as consequence of metabolic cooperation in a synthetic yeast community. Biotechnol J. 2016;11:1169–78. doi: 10.1002/biot.201500301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wargo MJ, Hogan DA. Fungal–bacterial interactions: a mixed bag of mingling microbes. Curr Opin Microbiol. 2006;9:359–64. doi: 10.1016/j.mib.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Embree M, Liu JK, Al-Bassam MM, Zengler K. Networks of energetic and metabolic interactions define dynamics in microbial communities. Proc Natl Acad Sci USA. 2015;112:15450–5. doi: 10.1073/pnas.1506034112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev. 1997;61:262–80. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helliwell KE, Wheeler GL, Leptos KC, Goldstein RE, Smith AG. Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol Biol Evol. 2011;28:2921–33. doi: 10.1093/molbev/msr124. [DOI] [PubMed] [Google Scholar]
- 18.Rodionova IA, Li X, Plymale AE, Motamedchaboki K, Konopka AE, Romine MF, et al. Genomic distribution of B-vitamin auxotrophy and uptake transporters in environmental bacteria from the Chloroflexi phylum. Environ Microbiol Rep. 2015;7:204–10. doi: 10.1111/1758-2229.12227. [DOI] [PubMed] [Google Scholar]
- 19.Hom EFY, Murray AW. Plant–fungal ecology. Niche engineering demonstrates a latent capacity for fungal–algal mutualism. Science. 2014;345:94–98. doi: 10.1126/science.1253320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Momeni B, Brileya KA, Fields MW, Shou W. Strong inter-population cooperation leads to partner intermixing in microbial communities. Elife. 2013;2:e00230. doi: 10.7554/eLife.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller MJI, Neugeboren BI, Nelson DR, Murray AW. Genetic drift opposes mutualism during spatial population expansion. Proc Natl Acad Sci USA. 2014;111:1037–42. doi: 10.1073/pnas.1313285111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratzke C, Gore J. Self-organized patchiness facilitates survival in a cooperatively growing Bacillus subtilis population. Nat Microbiol. 2016;1:16022. doi: 10.1038/nmicrobiol.2016.22. [DOI] [PubMed] [Google Scholar]
- 23.Carmona-Fontaine C, Deforet M, Akkari L, Thompson CB, Joyce JA, Xavier JB. Metabolic origins of spatial organization in the tumor microenvironment. Proc Natl Acad Sci USA. 2017;114:2934–9. doi: 10.1073/pnas.1700600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dietrich LEP, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC, Newman DK. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol. 2013;195:1371–80. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters AC, Wimpenny J. Oxygen profiles in, and in the agar beneath, colonies of Bacillus cereus, Staphylococcus albus and Escherichia coli. Microbiology. 1987;133:1257–63. doi: 10.1099/00221287-133-5-1257. [DOI] [PubMed] [Google Scholar]
- 26.Ratzke C, Gore J. Modifying and reacting to the environmental pH drives bacterial interactions. bioRxiv. 2017. 10.1101/136838. [DOI] [PMC free article] [PubMed]
- 27.Vylkova S. Environmental pH modulation by pathogenic fungi as a strategy to conquer the host Hogan DA (ed) PLoS Pathog. 2017;13:e1006149. doi: 10.1371/journal.ppat.1006149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gòdia F, Albiol J, Montesinos JL, Pérez J, Creus N, Cabello F, et al. MELISSA: a loop of interconnected bioreactors to develop life support in space. J Biotechnol. 2002;99:319–30. doi: 10.1016/S0168-1656(02)00222-5. [DOI] [PubMed] [Google Scholar]
- 29.Lucy M, Reed E, Glick BR. Applications of free living plant growth-promoting rhizobacteria. Antonie Van Leeuwenhoek. 2004;86:1–25. doi: 10.1023/B:ANTO.0000024903.10757.6e. [DOI] [PubMed] [Google Scholar]
- 30.Richardson AE, Lynch JP, Ryan PR, Delhaize E, Smith FA, Smith SE, et al. Plant and microbial strategies to improve the phosphorus efficiency of agriculture. Plant Soil. 2011;349:121–56. doi: 10.1007/s11104-011-0950-4. [DOI] [Google Scholar]
- 31.Weiss M, Waller F, Zuccaro A, Selosse MA. Sebacinales—one thousand and one interactions with land plants. New Phytol. 2016;211:20–40. doi: 10.1111/nph.13977. [DOI] [PubMed] [Google Scholar]
- 32.Mee MT, Wang HH. Engineering ecosystems and synthetic ecologies. Mol Biosyst. 2012;8:2470–83. doi: 10.1039/c2mb25133g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Widder S, Allen RJ, Pfeiffer T, Curtis TP, Wiuf C, Sloan WT, et al. Challenges in microbial ecology: building predictive understanding of community function and dynamics. ISME J. 2016;10:2557–68. doi: 10.1038/ismej.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherameti I, Tripathi S, Varma A, Oelmüller R. The root-colonizing endophyte Pirifomospora indica confers drought tolerance in Arabidopsis by stimulating the expression of drought stress-related genes in leaves. Mol Plant Microbe Interact. 2008;21:799–807. doi: 10.1094/MPMI-21-6-0799. [DOI] [PubMed] [Google Scholar]
- 35.Vadassery J, Ranf S, Drzewiecki C, Mithöfer A, Mazars C, Scheel D, et al. A cell wall extract from the endophytic fungus Piriformospora indica promotes growth of Arabidopsis seedlings and induces intracellular calcium elevation in roots. Plant J. 2009;59:193–206. doi: 10.1111/j.1365-313X.2009.03867.x. [DOI] [PubMed] [Google Scholar]
- 36.Waller F, Achatz B, Baltruschat H, Fodor J, Becker K, Fischer M, et al. The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc Natl Acad Sci USA. 2005;102:13386–91. doi: 10.1073/pnas.0504423102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Druege U, Baltruschat H, Franken P. Piriformospora indica promotes adventitious root formation in cuttings. Sci Hortic. 2007;112:422–6. doi: 10.1016/j.scienta.2007.01.018. [DOI] [Google Scholar]
- 38.Yadav V, Kumar M, Deep DK, Kumar H, Sharma R, Tripathi T, et al. A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. J Biol Chem. 2010;285:26532–44. doi: 10.1074/jbc.M110.111021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Kumar V, Sahai V, Bisaria VS. High-density spore production of Piriformospora indica, a plant growth-promoting endophyte, by optimization of nutritional and cultural parameters. Bioresour Technol. 2011;102:3169–75. doi: 10.1016/j.biortech.2010.10.116. [DOI] [PubMed] [Google Scholar]
- 40.Jacobs S, Zechmann B, Molitor A, Trujillo M, Petutschnig E, Lipka V, et al. Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol. 2011;156:726–40. doi: 10.1104/pp.111.176446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiang X, Weiss M, Kogel KH, Schäfer P. Piriformospora indica—a mutualistic basidiomycete with an exceptionally large plant host range. Mol Plant Pathol. 2011;13:508–518. doi: 10.1111/j.1364-3703.2011.00764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varma A, Sherameti I, Tripathi S, Prasad R, Das A. The symbiotic fungus Piriformospora indica: review. Fungal Assoc. 2012;130:39–254. [Google Scholar]
- 43.Zuccaro A, Lahrmann U, Güldener U, Langen G, Pfiffi S, Biedenkopf D, et al. Endophytic life strategies decoded by genome and transcriptome analyses of the mutualistic root symbiont Piriformospora indica. PLoS Pathog. 2011;7:e1002290. doi: 10.1371/journal.ppat.1002290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haas AL, Laun NP, Begley TP. Thi20, a remarkable enzyme from Saccharomyces cerevisiae with dual thiamin biosynthetic and degradation activities. Bioorg Chem. 2005;33:338–44. doi: 10.1016/j.bioorg.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 45.Hohmann S, Meacock PA. Thiamin metabolism and thiamin diphosphate-dependent enzymes in the yeast Saccharomyces cerevisiae: genetic regulation. Biochim Biophys Acta. 1998;1385:201–19. doi: 10.1016/S0167-4838(98)00069-7. [DOI] [PubMed] [Google Scholar]
- 46.Machado CR, Praekelt UM, de Oliveira RC, Barbosa AC, Byrne KL, Meacock PA, et al. Dual role for the yeast THI4 gene in thiamine biosynthesis and DNA damage tolerance. J Mol Biol. 1997;273:114–21. doi: 10.1006/jmbi.1997.1302. [DOI] [PubMed] [Google Scholar]
- 47.Wightman R. The THI5 gene family of Saccharomyces cerevisiae: distribution of homologues among the hemiascomycetes and functional redundancy in the aerobic biosynthesis of thiamin from pyridoxine. Microbiology. 2003;149:1447–60. doi: 10.1099/mic.0.26194-0. [DOI] [PubMed] [Google Scholar]
- 48.Kraft CE, Angert ER. Competition for vitamin B 1(thiamin) structures numerous ecological interactions. Q Rev Biol. 2017;92:151–68. doi: 10.1086/692168. [DOI] [PubMed] [Google Scholar]
- 49.Michal G, Schomburg D. Biochemical pathways: an atlas of biochemistry and molecular biology. Wiley, New York; 1999.
- 50.Begley TP, Downs DM, Ealick SE, McLafferty FW, Van Loon APGM, Taylor S, et al. Thiamin biosynthesis in prokaryotes. Arch Microbiol. 1999;171:293–300. doi: 10.1007/s002030050713. [DOI] [PubMed] [Google Scholar]
- 51.Jenkins AH, Schyns G, Potot S, Sun G, Begley TP. A new thiamin salvage pathway. Nat Chem Biol. 2007;3:492–7. doi: 10.1038/nchembio.2007.13. [DOI] [PubMed] [Google Scholar]
- 52.Jurgenson CT, Begley TP, Ealick SE. The structural and biochemical foundations of thiamin biosynthesis. Annu Rev Biochem. 2009;78:569–603. doi: 10.1146/annurev.biochem.78.072407.102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong HA, Khaneja R, Tam NMK, Cazzato A, Tan S, Urdaci M, et al. Bacillus subtilis isolated from the human gastrointestinal tract. Res Microbiol. 2009;160:134–43. doi: 10.1016/j.resmic.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 54.Mäder U, Schmeisky AG, Flórez LA, Stülke J. SubtiWiki—a comprehensive community resource for the model organism Bacillus subtilis. Nucleic Acids Res. 2012;40:D1278–87. doi: 10.1093/nar/gkr923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schyns G, Potot S, Geng Y, Barbosa TM, Henriques A, Perkins JB. Isolation and characterization of new thiamine-deregulated mutants of Bacillus subtilis. J Bacteriol. 2005;187:8127–36. doi: 10.1128/JB.187.23.8127-8136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castillo H, Aguilar C, Reyes CF, Morales GG. Biological control of root pathogens by plant-growth-promoting Bacillus spp.. In: Dr. Sonia Soloneski (Ed.), Weed and Pest Control - Conventional and New Challenges. InTech; 2013. 10.5772/54229.
- 57.Lu J, Frank EL. Rapid HPLC measurement of thiamine and its phosphate esters in whole blood. Clin Chem. 2008;54:901–6. doi: 10.1373/clinchem.2007.099077. [DOI] [PubMed] [Google Scholar]
- 58.Turner JT. Factors relating to peanut yield increases after seed treatment with Bacillus subtilis. Plant Dis. 1991;75:347. doi: 10.1094/PD-75-0347. [DOI] [Google Scholar]
- 59.Kempes CP, Okegbe C, Mears-Clarke Z, Follows MJ, Dietrich LEP. Morphological optimization for access to dual oxidants in biofilms. Proc Natl Acad Sci USA. 2014;111:208–13. doi: 10.1073/pnas.1315521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tacon FL, Skinner FA, Mosse B. Spore germination and hyphal growth of a vesicular–arbuscular mycorrhizal fungus, Glomus mosseae (Gerdemann and Trappe), under decreased oxygen and increased carbon dioxide concentration. Can J Microbiol. 1983;29:1280–5. doi: 10.1139/m83-200. [DOI] [Google Scholar]
- 61.Chatterjee A, Abeydeera ND, Bale S, Pai PJ, Dorrestein PC, Russell DH, et al. Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature. 2011;478:542–6. doi: 10.1038/nature10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalenitchenko D, Fagervold SK, Pruski AM, Vétion G, Yücel M, Le Bris N, et al. Temporal and spatial constraints on community assembly during microbial colonization of wood in seawater. ISME J. 2015;9:2657–70. doi: 10.1038/ismej.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ponomarova O, Gabrielli N, Sévin DC, Mülleder M, Zirngibl K, Bulyha K, et al. Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Syst. 2017;5:345–57. doi: 10.1016/j.cels.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Braakman R, Follows MJ, Chisholm SW. Metabolic evolution and the self-organization of ecosystems. Proc Natl Acad Sci USA. 2017;114:E3091–E3100. doi: 10.1073/pnas.1619573114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zerfaß C, Chen J, Soyer OS. Engineering microbial communities using thermodynamic principles and electrical interfaces. Curr Opin Biotechnol. 2017;50:121–7. doi: 10.1016/j.copbio.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 66.Großkopf T, Consuegra J, Gaffé J, Willison JC, Lenski RE, Soyer OS, et al. Metabolic modelling in a dynamic evolutionary framework predicts adaptive diversification of bacteria in a long-term evolution experiment. BMC Evol Biol. 2016;16:163. doi: 10.1186/s12862-016-0733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singhal U, Attri MK, Varma A. Mass cultivation of mycorrhiza-like fungus Piriformospora indica (Serendipita indica) by batch in bioreactor. In: Varma A., Prasad R., Tuteja N. (eds) Mycorrhiza - Function, Diversity, State of the Art. Cham: Springer International Publishing; 2017. p. 365–384.
- 68.Perotto S, Bonfante P. Bacterial associations with mycorrhizal fungi: close and distant friends in the rhizosphere. Trends Microbiol. 1997;5:496–501. doi: 10.1016/S0966-842X(97)01154-2. [DOI] [PubMed] [Google Scholar]
- 69.Pérez-Jaramillo JE, Mendes R, Raaijmakers JM. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol Biol. 2016;90:635–44. doi: 10.1007/s11103-015-0337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–38. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 71.Davison J. Plant beneficial bacteria. Biotechnology. 1988;6:282–286. [Google Scholar]
- 72.Veneault-Fourrey C, Martin F. 10 New Insights into Ectomycorrhizal Symbiosis Evolution and Function. In: Kempken F. (eds) Agricultural Applications. The Mycota (A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research), vol 11. Springer, Berlin, Heidelberg; 2013.
- 73.Vessey JK. Plant growth promoting rhizobacteria as biofertilizers. Plant and Soil. 2003;255:571–586. doi: 10.1023/A:1026037216893. [DOI] [Google Scholar]
- 74.Yurgel SN, Rice J, Domreis E, Lynch J, Sa N, Qamar Z, et al. Sinorhizobium meliloti flavin secretion and bacteria-host interaction: role of the bifunctional RibBA protein. Mol Plant Microbe Interact. 2014;27:437–45. doi: 10.1094/MPMI-11-13-0338-R. [DOI] [PubMed] [Google Scholar]
- 75.Lareen A, Burton F, Schäfer P. Plant root–microbe communication in shaping root microbiomes. Plant Mol Biol. 2016;90:575–87. doi: 10.1007/s11103-015-0417-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Veresoglou SD, Chen B, Rillig MC. Arbuscular mycorrhiza and soil nitrogen cycling. Soil Biol Biochem. 2012;46:53–62. doi: 10.1016/j.soilbio.2011.11.018. [DOI] [Google Scholar]
- 77.Fitter AH, Garbaye J. Interactions between mycorrhizal fungi and other soil organisms. Plant Soil. 1994;159:123–32. doi: 10.1007/BF00000101. [DOI] [Google Scholar]
- 78.Kohlmeier S, Smits THM, Ford RM, Keel C, Harms H, Wick LY. Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ Sci Technol. 2005;39:4640–6. doi: 10.1021/es047979z. [DOI] [PubMed] [Google Scholar]
- 79.diCenzo GC, Zamani M, Cowie A, Finan TM. Proline auxotrophy in Sinorhizobium meliloti results in a plant-specific symbiotic phenotype. Microbiology (Read, Engl) 2015;161:2341–51. doi: 10.1099/mic.0.000182. [DOI] [PubMed] [Google Scholar]
- 80.Nagae M, Parniske M, Kawaguchi M, Takeda N. The relationship between thiamine and two symbioses: root nodule symbiosis and arbuscular mycorrhiza. Plant Signal Behav. 2016;11:e1265723. doi: 10.1080/15592324.2016.1265723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Streit WR, Joseph CM, Phillips DA. Biotin and other water-soluble vitamins are key growth factors for alfalfa root colonization by Rhizobium meliloti 1021. Mol Plant Microbe Interact. 1996;9:330–8. doi: 10.1094/MPMI-9-0330. [DOI] [PubMed] [Google Scholar]
- 82.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Altschul S. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Federhen S. The NCBI Taxonomy database. Nucleic Acids Res. 2012;40:D136–43. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 90.Pieterse, CMJ, de Jonge, R, Berendsen, RL The Soil-Borne Supremacy. Trends in Plant Science. 2016; 21 (3):171-173 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information (containing Supplementary Figures and Tables)(PDF 6688 kb)