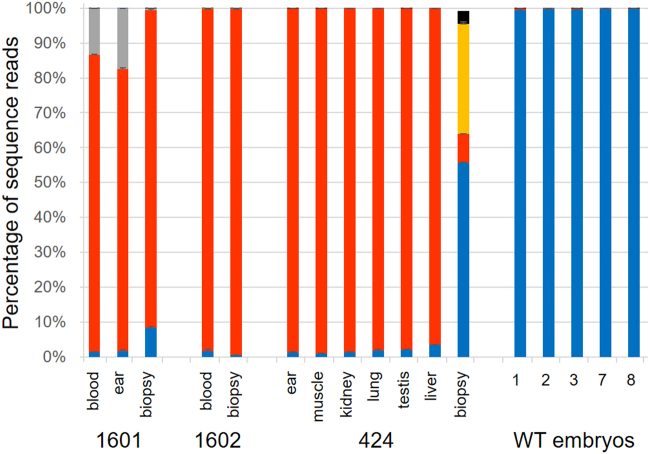
Figure 4.
Deep sequencing analysis of the edited LGB locus in the calves 1601, 1602 and 424 and corresponding embryo biopsies. DNA isolated from blood, ear fibroblasts (ear), a selection of tissues (as indicated) and embryo biopsies (biopsy) were analyzed for the presence of TALEN-mediated edits and unaltered wild type sequences of the LGB target locus by enumerating corresponding sequencing reads for 1601, 1602, 424 associated samples in addition two five wild type embryo samples (1, 2, 3, 7, 8). The bars depict the percentage of all joined paired end sequence reads in the indicated samples for the HDR-generated nine bp deletion allele (red), wild type allele (blue) and additional 21 bp (grey), 24 bp (yellow) and 66 bp (black) deletion alleles that were identified by the deep sequencing analysis. The values that were used to generate the graph can be found in Supplementary Table S2. Error bars indicate standard errors.