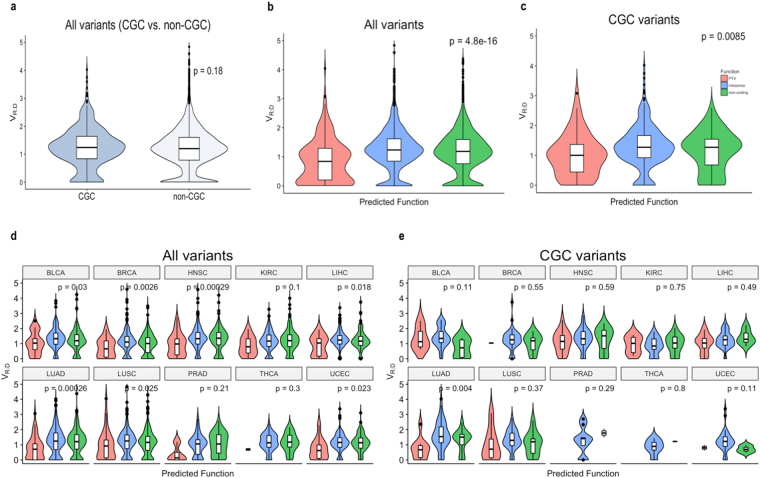
Figure 2.
(a) Distribution of VR:D in somatic mutations between CGC and non-CGC genes. (b) Distribution of VR:D in somatic mutations categorized based on their predicted effect on the protein function in the entire dataset and in the CGC subset. (c) The mutation categories correspond to: generating a premature termination codon variant (PTV), substitution of an amino acid (missense) or not altering a coding sequence (silent). In both analyses the PTVs presented with significantly different distribution of VR:D (p < 0.05) expressed as lower average allele fraction and a higher proportion of SOM-L mutations (VR:D ~ 0). However, while still significantly different, the PTV VR:D in the CGC genes appears more similar to the VR:D of missense and silent variants, as compared to the pooled data from all genes. (d) Distribution of VR:D in PTV, missense and silent variants in individual cancer types across the entire dataset, and (e) in CGC-genes.