Figure 3. NMR based mutagenesis refines the surface of the ITK PHTH domain that contacts the ITK kinase domain.
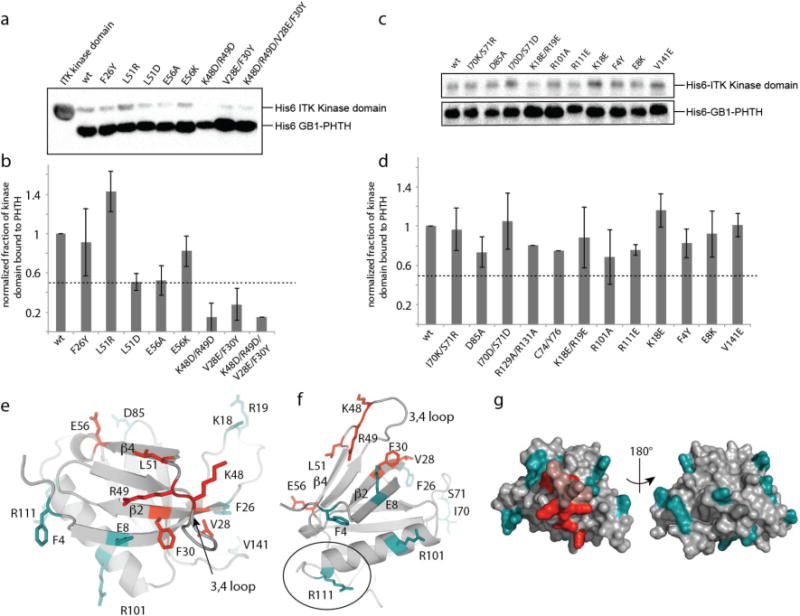
(a,c) Anti-His6 blots showing ITK kinase domain (4μM) associates to different extents with wild type and mutant ITK PHTH domain (2μM) that is immobilized on IgG sepharose beads. (b,d) The fraction of ITK kinase domain bound to wild type and mutant ITK PHTH domains; bound kinase is normalized to His6-GB1 PHTH level in each lane and then compared to wild type (wt). The dotted line shows a decrease in ITK PHTH/kinase domain interaction of 50%. Data shown in (a) and (c) are representative of three independent experiments. (e,f) The binding surface determined from the data in (a) and (c) is mapped onto the ITK PHTH domain computational model, labeled and shown in red. The interface residues are located on the β4 and β2 strands as well as the β3-β4 loop. Those residues that, upon mutation, do not disrupt the ITK PHTH/kinase domain interaction are labeled and shown in cyan. In (f) R111 is circled as it corresponds to the BTK residue R133 that is at the interface of the tethered BTK PHTH and kinase domains for which a crystal structure was recently solved15. (g) Two views of the ITK PHTH surface residues shown in (e) and (f) where red indicates involvement in the ITK PHTH/kinase interaction and cyan corresponds to those residues for which mutation does not affect the ITK PHTH/kinase interaction. The β3-β4 loop residues that are within the binding site identified here are colored pink but were not probed directly by mutagenesis.
