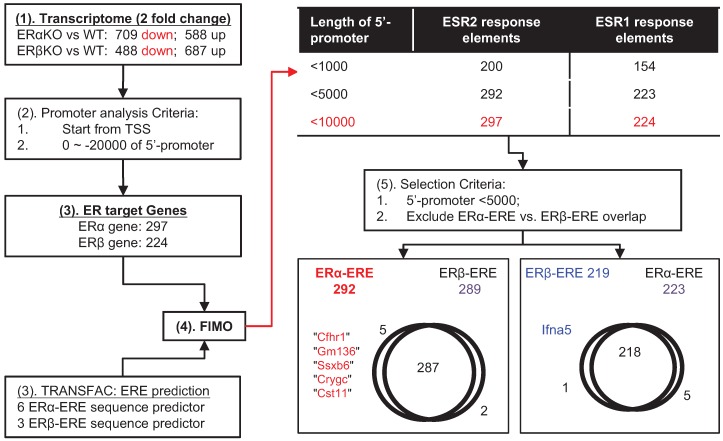
Figure 3.
The target ERα- and ERβ-related genes predicted by a bioinformatics approach. (1) The non-overlapping downregulated genes (i.e., genes encoding transcription factors) in the ERα-KO or ERβ-KO mouse transcriptome were chosen for analysis. (2) The genes were subjected to promoter analysis (to locate promoter sequences within ~ -20 kb upstream from the transcription start-site). (3) Both R-software and TRANSFAC were used to analyze EREs (specific for ERα or ERβ; Figure S2) and found 297 genes for ERα and 224 genes for ERβ. (4) FIMO was used to locate the promoter sequences of ERα-ERE and ERβ-ERE, and most EREs were located within -5000 bp of their promoters. (5) The ERα-ERE vs. ERβ-ERE region of overlap was excluded to distinguish ERα-specific from ERβ-specific target genes. The final result identified Cfhr1, Gm136, Ssx6, Cryc, and Cst11 as ERα-specific and Ifna5 as ERβ-specific.