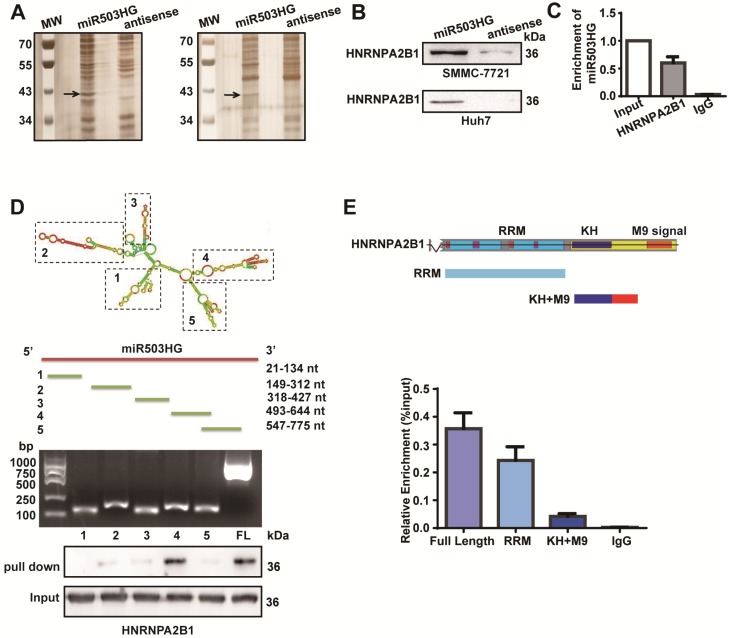
Figure 3.
miR503HG physically interacts with HNRNPA2B1 in HCC cells. (A) RNA pull-down assays were performed in SMMC-7721 cells twice. Biotinylated miR503HG (left lane) or antisense RNA (right lane) was incubated with cell extracts and targeted with streptavidin beads; the associated proteins were resolved on a gel. The highlighted region was submitted for mass spectrometry. (B) Western blotting analysis of the specific association of HNRNPA2B1 with miR503HG in SMMC-7721 and Huh7 cells. (C) RIP enrichment was determined as RNA associated with the HNRNPA2B1 IP relative to an input control. The data represent the average and standard deviation of three independent experiments. (D) Deletion mapping of the HNRNPA2B1-binding domain in miR503HG. Top: graphic illustration of predicted miR503HG secondary structure (LNCipedia, http://www.lncipedia.org), and the truncation of miR503HG according to the stem-loop structure. Middle: the in vitro transcribed full-length and deletion fragments of miR503HG. Bottom: Western blotting analysis of HNRNPA2B1 in protein samples pulled down by the different miR503HG constructs. (E) Deletion mapping of the miR503HG-binding domain in HNRNPA2B1. Top: diagrams of full-length HNRNPA2B1 and the domain-truncated fragments RNA recognition motif (RRM), hnRNP K homology domain (KH), M9 signal (MN). Bottom: qPCR analysis of HNRNPA2B1 retrieved by full-length or domain-truncated HNRNPA2B1-HA using a HA antibody. RIP assays were performed using Huh7 cells transfected with the indicated vectors.