Figure 2. Validation of predicted PA genes using multiple approaches.
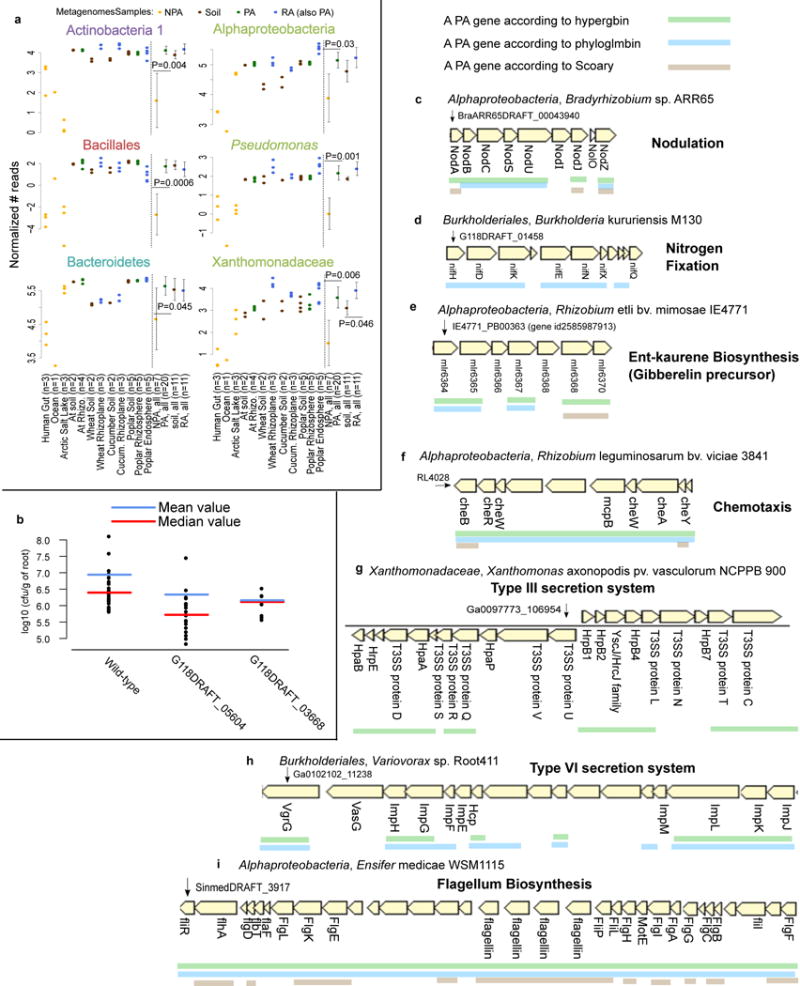
a. PA genes, which were predicted based on isolate genomes, are more abundant in PA metagenomes than in NPA metagenomes. Reads from 38 shotgun metagenome samples were mapped to significant PA, NPA, RA, and soil genes predicted by Scoary. P values are indicated for the significant differences between the PA and NPA or RA and soil in each taxon (two sided t-test). Full results and explanation for normalization are presented in Supplementary Figure 14. b. Rice root colonization experiment using wild type Paraburkholderia kururiensis M130 or knockout mutants for two predicted PA genes. Two mutants exhibited reduced colonization in comparison to wild type: G118DRAFT_05604 (q-value = 0.00013, wilcoxon rank sum test) encodes an outer membrane efflux transporter from the nodT family, and G118DRAFT_03668 (q-value = 0.0952, wilcoxon rank sum test), a Tir chaperone protein (CesT). Each point represents the average count of a minimum of 3-6 plates derived from the same plantlet, expressed as cfu/g of root. c-i. Examples of known functional PA operons captured by different statistical approaches. The PA genes are underlined. c. Nod genes, d. NIF genes, e. ent-kaurene (gibberelin precursor), f. Chemotaxis proteins in bacteria from different taxa. g. Type III secretion system. h. Type VI secretion system, including the imp genes (impaired in nodulation), i. Flagellum biosynthesis in Alphaproteobacteria. Below each gene appears the gene symbol or the protein name where such information was available.
