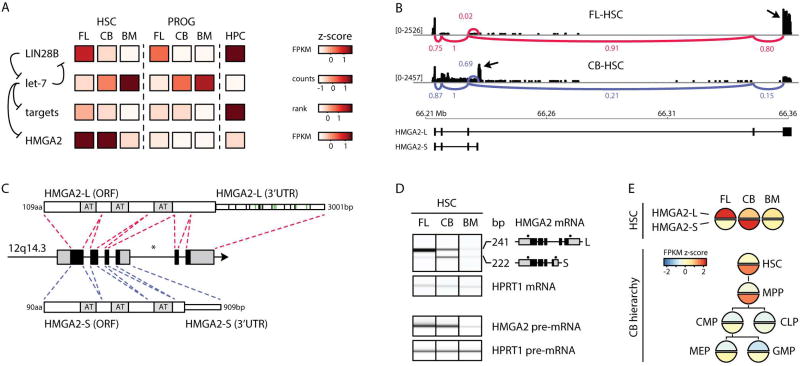
FIGURE 2. HMGA2 alternative splicing in human HSCs.
A- Heatmap showing expression of LIN28B, let-7 family members, the ranked median expression of the top 100 predicted let-7 targets, and HMGA2. Previously dissected regulatory relationships of the LIN28-let-7-HMGA2 pathway (Viswanathan et al., 2008) are indicated with arrows. Scales are shown on the right as normalized z-scores.
B- Visualization of RNA-seq reads (black bars) mapping at HMGA2 (drawn to scale) in the indicated samples. A sashimi plot displaying the major splice junctions (FL-HSCs, red; CB-HSCs, blue) is superimposed. The abundance of each splicing junction is indicated and is normalized to the counts for the shared exon 2–3 splice junction (with value of 1). Arrows indicate the reads mapping to the terminal exons that distinguish the HMGA2-L and HMGA2-S isoforms.
C- Structure of the human HMGA2 locus (not drawn to scale). In the middle, locus coordinates are indicated along with coding exons (black) and UTRs (gray). Asterisk indicates location of major chromosomal rearrangements detected in malignancies (Kazmierczak et al., 1996). Red and blue dashed lines indicate how exons are spliced to result in HMGA2-L (top) and HMGA2-S (bottom). Gray boxes indicate the AT-binding hook domains. Green and black slashes indicate predicted binding sites of let-7 family members and other conserved HSC-expressed miRNAs respectively.
D- (top) Digital gel from RT-PCR electropherogram showing expression of HMGA2-L and HMGA2-S from the indicated HSC populations. Sizes of the amplicons are indicated on the right, along with the structure of each isoform (coding exons in black, UTRs in grey) and the position of the primers used for PCR co-amplification (black dots). (bottom) Digital gel from RT-PCR electropherogram showing expression of HMGA2 pre-mRNA from the indicated HSC populations. HPRT1 pre-mRNA was used as the control for both gels. Expression levels of HMGA2 isoforms are indicated in Figure S2 and Table S2A.
E- Shaded circles show HMGA2 isoform expression (z-score normalized FPKM values) across HSCs and CB hierarchy from publicly available data (Stunnenberg et al., 2016). MPP: multipotent progenitors; CMP: common myeloid progenitors; MEP: myeloid erythroid progenitors; GMP: granulocyte macrophage progenitors; CLP: common lymphoid progenitors.