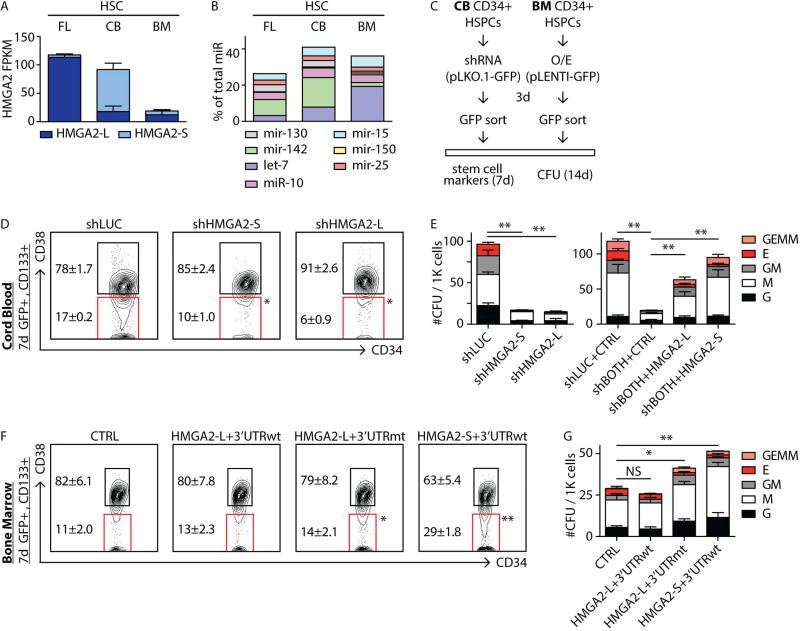
FIGURE 5. Modulation of HMGA2 isoforms in human HSPCs.
A,B- Absolute expression of the indicated HMGA2 isoforms (in FPKM) and miRNA families (as percentage of total measured miRNA content) in the indicated HSC populations.
C- Scheme of HMGA2 isoform modulation experiments in CB and BM CD34+ hematopoietic stem and progenitor cells (HSPCs). Lentiviral constructs (pLKO.1 and pLENTI) also express GFP as a marker to allow for isolation of infected cells.
D- Phenotypic analysis of the stem cell compartment (CD133+CD34+CD38−) in CB CD34+ cells transduced with negative-control hairpin (luciferase - shLUC) or HMGA2 isoform-specific shRNAs (shHMGA2-S or shHMGA2-L) following 7 days in culture.
E- (left) Clonogenic progenitor assay of CD34+ CB cells following knockdown of HMGA2 isoforms. Cells transduced with shLUC, shHMGA2-S and shHMGA2-L hairpins were plated 3 days post-infection and CFU potential was measured 14 days post-plating. (right) CFU potential of CD34+ CB transduced with hairpins against both HMGA2 isoforms (shBOTH) was rescued upon overexpression of either the HMGA2-L or HMGA2-S ORFs.
F- Phenotypic analysis of the stem cell compartment (CD133+CD34+CD38−) in BM CD34+ cells transduced with control (CTRL), HMGA2-L+3’UTRwt, HMGA2-L+3’UTRmt, or HMGA2-S+3’UTRwt constructs after 7 days in culture.
G- Clonogenic progenitor assay of CD34+ BM cells following overexpression of HMGA2 isoforms. Cells transduced with control (CTRL) were plated 3 days after infection and CFU potential was measured 14 days post-plating. Mean +/− s.e.m. values are shown for D, E, F, G. Analysis of deviance for generalized linear models (CFU analyses) or repeated measures ANOVA (FACS analyses) were used, p-values: <0.05 (*) or <0.01 (**), non-significant (NS).