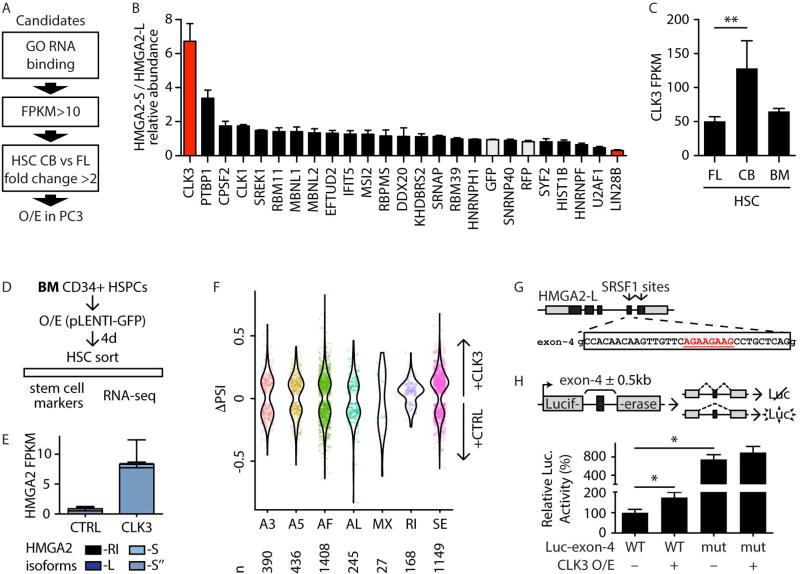
FIGURE 6. CLK3 affects HMGA2 splicing through SRSF1.
A- Schematic representation of the filtering strategy to identify regulators of HMGA2 splicing. Genes were selected for the GO term “RNA binding” (GO:0003723), minimum expression, and ratio of expression in CB-HSC versus FL-HSC, as in the scheme.
B- Ratio of expression of HMGA2-S/HMGA2-L following overexpression of the indicated candidate splicing factors in PC-3 cells. Controls (GFP and RFP) are shaded in gray. Values were normalized to the HMGA2-S/HMGA2-L ratio upon control GFP overexpression and shown as mean +/− s.e.m.
C- CLK3 expression by RNA-seq (in FPKM) in the indicated HSC populations. Mean +/− s.d. values are shown. FDR<0.01 (**).
D- Schematic representation of CLK3 and HMGA2 modulation in BM HSPCs.
E- HMGA2 isoform quantification by RNA-seq (in FPKM) in BM-HSCs isolated upon overexpression of CLK3 or control (CTRL) four days post-infection. Mean +/− s.d. values are shown. PCR validation is shown in Figure S5E.
F- Violin plot representing distributions of statistically significant (p<0.05) ΔPSI values for different classes of PSI events (as in Figure 1H) in CLK3 overexpression as compared to vector control. Individual significant events are shown. The number of events of each PSI class are shown below the plot.
G- Schematic representation of HMGA2-L genomic structure (coding exons in black, UTRs in grey). SRSF1 binding motif sites are indicated by arrows in the corresponding exons. Sequence of exon 4 is indicated, and the predicted SRSF1 site (underlined in red) is shown.
H- Schematic representation of the luciferase-based splicing reporter constructs. The pcDNA3.1-Luc plasmid contains an intron-spaced Firefly luciferase coding sequence (grey boxes) that produces bioactive luciferase protein only upon proper splicing of the intervening intron. The HMGA2-L genomic region encompassing exon 4 (black box) plus 500 bp on either side of the flanking introns (wild-type [WT] or mutated [mut] by deletion of SRSF1 site) was cloned into the intron of pcDNA3.1-Luc. Its inclusion in the luciferase coding sequencing abolishes bioactive luciferase. Normalized luciferase activities were tested with or without CLK3 overexpression and reported with respect to Luc-exon-4 WT set to 100%. Mean +/− s.e.m. values are shown. Repeated measures ANOVA was used, p-value <0.05 (*).