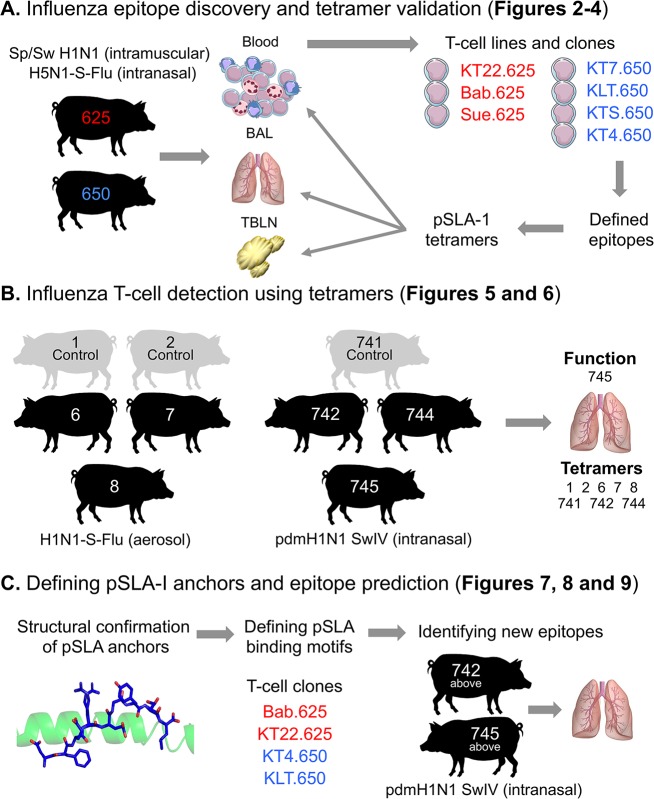
Fig 1. Study overview.
The inbred Babraham pig was used throughout this study for vaccination and infection, with each pig assigned an identifying number, shown here within each silhouette. (A) Pigs 625 (red) and 650 (blue) were vaccinated intranasally and intramuscularly as depicted. Blood, bronchoalveolar lavage (BAL) and tracheobronchial lymph nodes (TBLNs) were harvested, with peripheral blood mononuclear cells (PBMCs) purified from blood and single suspensions from BAL and TBLNs generated for experiments. Overlapping peptides from the NP of PR8 were used to create T-cell lines (Fig 2) and T-cell clones (named and shown in red or blue text) (Fig 3). The red clones came from pig 625 (red) and the blue from pig 650 (blue). The clones were used to define minimal NP peptides, which were subsequently refolded with SLA-1*14:02 or SLA-2*11:04 to create pSLA-I tetramers. The tetramers were used to stain the clones (Fig 3) and harvested tissues from pigs 625 and 650 (Fig 4). (B) The BAL from pigs vaccinated or infected intranasally with influenza, as shown, were stained with the tetramers from A (Figs 5 and 6). The BAL from pig 745 was used for ex vivo ELISPOTS. (C) SLA-1*14:02 or SLA-2*11:04 were refolded with the epitopes defined in A to confirm peptide anchor residues (Fig 7). T-cell clones from A were used to define a SLA-1*14:02 or SLA-2*11:04 peptide anchor binding motif (Fig 8), which were then used to predict other influenza epitopes, tested using BAL from the two pigs shown (Fig 9).