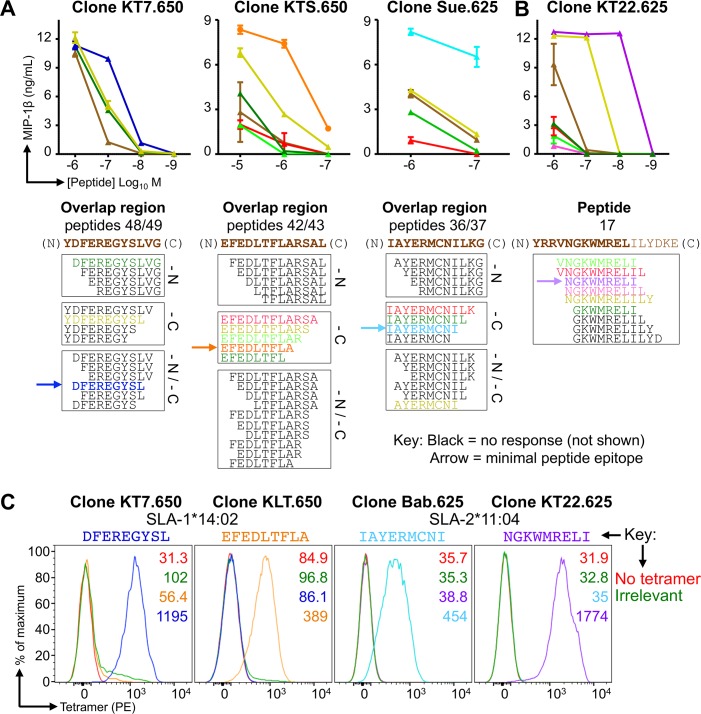
Fig 3. Minimal epitope identification and pSLA tetramer staining of influenza-specific T-cell clones.
CD8β clones were grown from T-cell lines generated from Babraham pigs 625 and 650 using overlapping peptides from the NP of S-FLU (PR8) (Fig 2). The .650 or .625 indicates the pig the clones were grown from. (A) Clones KT7.650, KTS.650 and Sue.625 responded to the overlapping peptide regions shown in brown text below each graph. Clones were incubated with decreasing concentrations (upper panel) of truncated versions of the respective peptide (sequences shown in the lower panel). Truncations were performed at the amino (N) and/or carboxyl (C) terminal ends of each peptide as indicated (lower panel). MIP-1β ELISAs were performed to assess T-cell activation after overnight incubation. Peptides that did not elicit a repsonse are shown in black and are not displayed on the graphs. The minimal peptide epitope is indicated by the arrow. (B) Using a similar approach as in (A). In order to induce maximal activation and define the minimal epitope, clone KT22.625 (right) required the isoleucine from peptide 17, which was not present in the overlap region of 16/17. (C) The minimal epitopes defined in (A and B) were refolded with both SLA-1*14:02 and SLA-2*11:04, with successful refolding determining restriction. pSLA-I tetramers were assembled and used to stain clones with respective peptide specificity, as shown. Irrelevant tetramers: SLA-1*14:02-AFAAAAAAL and SLA-2*11:04-AGAAAAAAI.