Abstract
A bacterial artificial chromosome (BAC) library for G. mustelinum Miers ex G. Watt (AD4) was constructed. Intact nuclei from G. mustelinum (AD4) were used to isolate high molecular weight DNA, which was partially cleaved with Hind III and cloned into pSMART BAC (Hind III) vectors. The BAC library consisted of 208,182 clones arrayed in 542 384-microtiter plates, with an average insert size of 121.72 kb ranging from 100 to 150 kb. About 2% of the clones did not contain inserts. Based on an estimated genome size of 2372 Mb for G. mustelinum, the BAC library was estimated to have a total coverage of 10.50 × genome equivalents. The high capacity library of G. mustelinum will serve as a giant gene resource for map-based cloning of quantitative trait loci or genes associated with important agronomic traits or resistance to Verticillium wilt, physical mapping and comparative genome analysis.
Introduction
Gossypium genus comprises more than 50 species, including eight diploid groups and six tetraploid species [1–3]. Among the six tetraploid species, G. hirsutum L. (AD1) and G. barbadense L. (AD2) have been domesticated. The remaining tetraploid species are all wild species, and have much narrower distributions: G. tomentosum Nuttall ex Seemann (AD3) in the Hawaiian Islands, and G. mustelinum Miers ex Watt (AD4) in NE Brazil, G. darwinii Watt (AD5) in the Galapagos Islands[4], except for G. ekmanianum Wittmack (AD6), whose distribution needs to be further determined[3, 5]. Because of long-term natural selection under the distrubution environment, these wild species have rich genetic diversity and contain many excellent genes which can be used in cotton breeding, such as drought resistance, disease resistance, cold resistance, and the potential properties of fine fibers [6]. Phylogenetic and phenetic ananlyses demonstrate that G. mustelinum had been isolated as one branch of the earliest split following allopolyploid formation, and is genetically farthest from G. hirsutum and other tetraploid species [4, 5, 7]. So most likely there have some new genetic loci that can be used for genetic improvement of upland cotton, which is helpful to solve the serious bottleneck of genetic diversity in the development of upland cotton varieties [8–10]. Moreover, G. mustelinum showed a special chromosome structure based on the study of molecular cytogenetics [11]. As a very special tetraploid wild cotton species, G. mustelinum has irreplaceable value in basic research and germplasm. However, there are few reports on its genome research because of the difficulty to obtain experimental material and complexity of genome [12].
Bacterial artificial chromosome (BAC) libraries carrying large insert genomic DNA, are essential genomic resources for map-based gene cloning and and physical map construction [13–15], development of markers based on BAC-end sequences [16, 17], genome sequencing of complex genomes [18]. So far, dozens of BAC libraries have been reported for different Gossypium species. Such as libraries from G. hirsutum [19–23], G. barbadense [24], G. tomentosum [25], G. arboretum [26], G. herbaceum var. africanum [27], and G. raimondii [28]. Based on these resources, researches on evolutionary relationship of Gossypium [29], individual chromosome identification and naming [30, 31], physical map construction and guidance in genome sequence assembly [32, 33] have been carried out.
In this paper, we firstly report the construction of a high capacity library in G. mustelinum, a wild tetraploid species, which is far from G. hirsutum in phylogeny, and has many excellent traits, including finer fibers and Verticillium resistance, which will help to save cotton germplasm resources, and provide important genome resources for cultivated cotton breeding programs about genetic diversity.
Materials and methods
Plant material and high-molecular-weight (HMW) DNA preparation
Wild tetraploid cotton species, G. mustelinum (AD4) (accession P0811704) were conserved in the greenhouse at CRI-CAAS in Anyang City, Henan Province, China. Cotton mature plants were grown in dark for about 10 day to prepare etiolated young leaf tissues. For chromosome preparation, seeds of G. mustelinum were planted in nutrition bowls for 7 d to harvest root tip.
The etiolated young leaf tissues were sampled, then immediately frozen in liquid nitrogen. Nuclei were isolated from about 30 g etiolated young leaves, according to the procedure described by Zhang et al. [34] and Wang et al. [24]. Purified nuclei were embedded in 1% low melting point (LMP) agarose to make agarose plugs. After lysis with proteinase K, the quality of HMW DNA was tested by pulse-field gel electrophoresis (PFGE) at 6 V/cm, switch times of N/S 50 s, E/W 50 s, 11°C for 18 h.
Generation of HMW genomic DNA fragments for cloning
The HMW DNA was digested with different Hind III (TaKaRa) concentrations to determine the optimum partial restriction digestion conditions for generating the highest percentage of DNA fragments between 100 and 300 kb. Every one half of the three randomly selected DNA plugs was equilibrated with restriction enzyme buffer for 30 min, then digested respectively with 0, 1, 2, 3, 4, 5, and 6 U Hind III in 37°C water bath for 30 min. The digested products were tested by PFGE at 6 V/cm, switch times of N/S 50 s, E/W 50s, 11°C for 18 h. Based on the optimal digestion conditions, digestion of the nucleus DNA were carried out. Then, two rounds of size selection of the partially digested products were carried out. The first size selection was aimed to recover gel fraction containing 100–300 kb fragments by PFGE at 6 V/cm, switch times of N/S 50 s, E/W 50s, 11°C for 18 h. The second size selection was performed using the first size-selected fractions cut from the center part of the gel fractions corresponding to 100–200 kb fragments and 200–300 kb fragments by PFGE at 6 V/cm, switch times of N/S 4 s, E/W 4s, 11°C for 14 h. The size-selected DNA was electroeluted in BIO-RAD PowerPac Electro-Eluter at 10 mA for 2.5 h. The concentrations of the electroeluted DNA were measured by 1% agarose gel electrophoresis (85 V, 40 min) using standard λDNA with different concentration gradient.
Ligation and bacterial transformations
The pSMARTBAC (Hind III) cloning vector (Lucigen) was used for library construction. Electroeluted DNA fragments and vector (molar ratio 1:10) were ligated using T4 DNA ligase in a 50 μL reaction solution at 16°C for 16 h. Then, the reaction solutions were heated to 65°C for 10 min to inactivate ligase, nextly, were desalted and concentrated by floating on ddH2O/PEG8000 with 0.025 μm Millipore filters for 2 h/1 h respectively.
Total ligation product (about 1–2 μL) was transformed into 20 μL EPI300 cells (Epicentre) using a MicroPulser Electroporation Apparatus (Bio-Rad) with settings of 2.5 kV, charge time 3 ms. Next, transferred cells culturing, positive clones picking and storing were performed according to the protocol described by Wang et al. [24].
BAC clones characterization
To estimate insert sizes, BAC clones were selected at random from the library and inoculated into 8 mL 2 × YT medium containing chloramphenicol (12.5 μg/mL) and incubated at 37°C for 14 h. BAC-DNA was isolated using Hipure BAC Mini Kit (Magen) according to the handbook. BAC-DNA (10 μL) was digested with 3 U of Not I enzyme for 3 h at 37°C. Insert sizes were estimated based on results of PFGE with 𝜆 PFG Ladder (NEB) as a reference.
Library stability was tested using 5 BAC clones selected at random from the library according to the protocol described by Hu et al. [26].
BAC library screening and positive BAC clone FISH verifying
To evaluate the BAC library, the SSR marker Gh216, once used to obtain a subgenome-specific BAC clone[33] was selected to screen the library using bacteria liquid-PCR according to our protocol previously described [15]. The primers (F/R) of Gh216 (TCCACATTCCCATGCACTACTC/CTAAAACCTTATACATACAAAATGCAGC) were synthesized by Shanghai Sangon Biotech Inc. The selected positive BAC clones were cultivated with LB medium containing 12.5 μg/ml chloramphenicol for 14 h. Then BAC-DNA were isolated using Hipure BAC Mini Kit (Magen) according to the handbook and used to label probes for FISH. Chromosome preparation and FISH were carried out following previous protocol [15].
Results
High-molecular-weight genomic DNA preparation
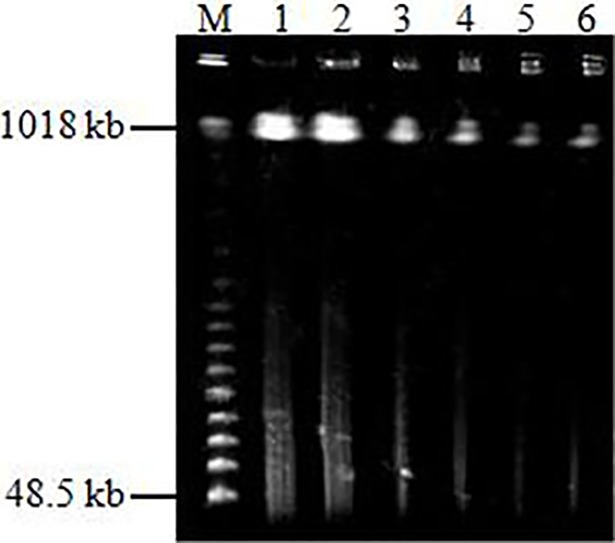
Undigested HMW DNAs isolated from etiolated young leaves had a mean length 1000 kb, and were seldom degraded (Fig 1), which can meet the needs of library construction.
Fig 1. The quality of HMW DNA tested by PFGE.
M, 𝜆 PFG Ladder (NEB); Lanes 1–6 were 1/2, 1/2, 1/4, 1/4, 1/8, 1/8 plug DNA.
The optimum partial restriction digestion condition and insert DNA prepration
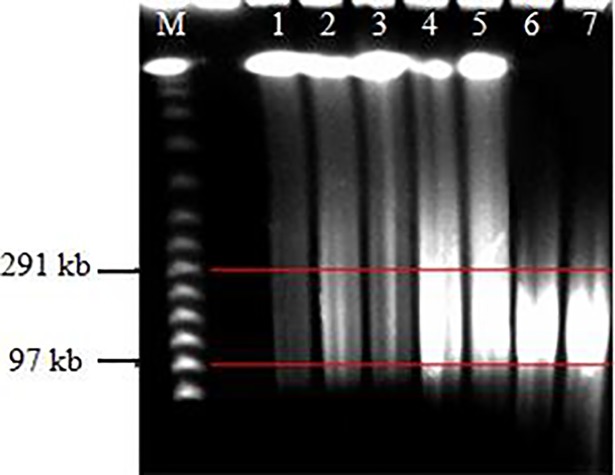
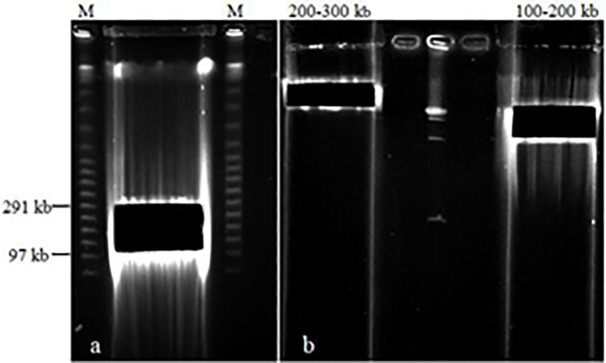
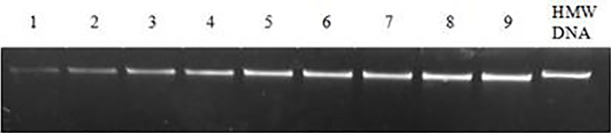
The suitable insert fragment length for BAC library construction is 100–300 kb. Digestion time and enzyme concentration are key factors for size control. Based on the result of PFGE, the optimal condition was digestion with 3 U Hind III at 37°C for 30 min, which concentrated more fragments between at 100–300 kb (Fig 2). A mass digestion was performed under the above condition to produce insert DNA. The digested DNA was treated with two rounds of electrophoresis, the fragments of 100–200 kb and 200–300 kb were recovered from the gel (Fig 3). The concentrations of the electroeluted DNA was approximately 5 ng/μL based on the detection with concentration-knownλDNA (Fig 4), which suits for the needs of connection transformation.
Fig 2. Determination of the optimized enzyme dosage for HMW DNA.
M, 𝜆 PFG Ladder (NEB); 1–7 indicated 0 U, 1 U, 2 U, 3 U, 4 U, 5 U, 6 U of Hind III, respectively.
Fig 3.
The first selection (a) and the second selection (b) restriction fragment of DNA.
Fig 4. Detection of HMW DNA concentrations.
Lanes 10 was HMW DNA; Lanes 1–9, λDNA concentrations were 1 ng/μL, 2 ng/μL, 3 ng/μL, 4 ng/μL, 5 ng/μL, 6 ng/μL, 7 ng/μL, 8 ng/μL, 10 ng/μL, respectively.
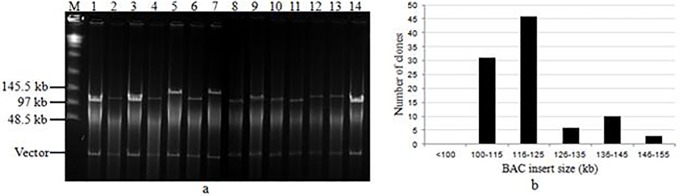
BAC library construction and characterization
The library was constructed after ligation and electroporation, which consists of 208,182 individual clones. All of the clones were handpicked and stored in 542 384-microtiter plates (Table 1). Ninty-eight BACs were randomly selected from the BAC library for size estimation. After digestion of BAC-DNA with Not I enzyme, the insert DNA fragment was separated from the vector (Fig 5A). Based on analysis, the average insert size of the BAC library was 121.72 kb ranging from 100 to 150 kb (Fig 5B). About 2% of the clones did not contain inserts (2 enpty clones to 98 selected clones). Based on an estimated genome size of 2372 Mb for G. mustelinum [35], the BAC library was estimated to have a total coverage of 10.50 × genome equivalents. The probability of finding any specific locus from this BAC library is estimated to be 99.9% according to the formula N = ln(1-p)/ln(1-f/G), where P is the probability, N is the number of clones, f is the average insert size of clones, and G is the haploid genome size [36].
Table 1. Statistics of BAC library of G. mustelinum.
| Clone numbers | 208,182 |
| Vector | pSMARTBAC (Hind III) |
| Comptetent cell | EPI300 cells |
| Average inset size | 122 kb |
| Haploid genome equivalent | 10.50 × |
| Insert-empty BACs | 4,164 (2%) |
Fig 5. Evaluation of insert sizes of BAC clones.
a, M, 𝜆 PFG Ladder (NEB); Lanes 1–14, NotI enzyme digestion of BAC single clones. b, insert size distribution.
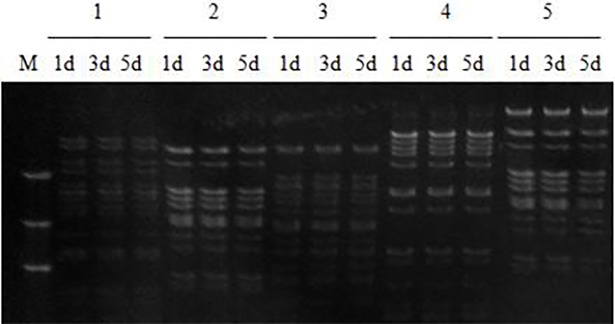
We analyzed the Hind III restriction patterns of five BAC clones after culturation of 1 d, 3 d and 5 d. No visible changes were seen in fingerprints of three BAC-DNA samples of each BAC clone (Fig 6), indicating the good stability of the BAC library.
Fig 6. Stability analysis of five BAC clones.
M, DL2000 DNA Marker; 1–5, five BAC single clones; 1d, 3d, 5d, samples from the first day, the third day and the fifth day.
BAC library screening and FISH verifying
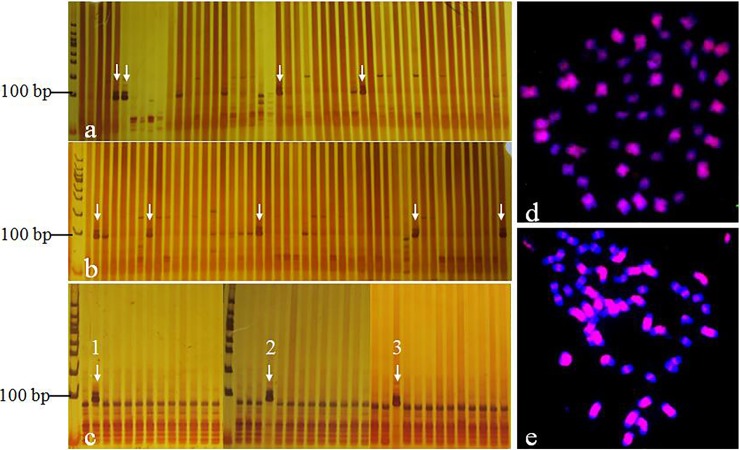
For library screening, we constructed 96 super-pools containing 36,864 BAC clones. After bacteria liquid-PCR screening with Gh216 SSR primers, nine super-pools with positive bands were obtained (Fig 7A and 7B). Further PCR screening against corresponding row and column pools, three positive BAC clones for Gh216 SSR marker were identified finally (Fig 7C, Table 2). According to the formula N = ln(1-p)/ln(1-f/G), the probability of finding a specific locus from a set of 36,864 BAC clones of G. mustelinum is about 85%, which is less than the success rate of three to one. This result was consistent with previous studies about Gh216-anchored BAC clone 57I23, which enriched in repeats distinguishing subgenome of allotetraploidy cottons [33]. In order to verify genome distribution of the identified BAC clones, FISH was performed using BAC-DNA of 48M15 as probe to hybridized with metaphase chromosomes G. mustelinum. Dispersed signals on the chromosomes were viewed (Fig 7D), but with a little lower coverage relative to BAC 57I23 identified previously (Fig 7E).
Fig 7. Positive BAC clones and FISH mapping.
a and b, nine positive super-pools (white arrows) based PCR-screening of 96 super-pools; c, three positive BAC clones (white arrows, 1-32N22, 2-48M15, 3-89H09) from 3 of 9 super-pools; d, FISH maping of 48M15 (red) on metaphase chromosomes of G. mustelinum; e, FISH maping of 57I23 (red) on metaphase chromosomes of G. mustelinum.
Table 2. BAC clones screened from the BAC library.
| SSR marker | Screened BAC clones |
|---|---|
| Gh216 | 32N22, 48M15, 89H09 |
Discussion
The obtaintion of high-quality nuclear DNA is the foundation for construction of BAC library. Generally, yellowing cotyledon of seedlings were selected as the materials for DNA extraction to avoid contamination of the chloroplast DNA. As rare wild resources, G. mustelinum has fewer seeds and lower seed germination rate, yellowing cotyledons are not easy to obtain. In this paper, we chose mature plants growing perennially in greenhouse to prepare etiolated young leaf tissues by growing in dark for about 10–15 days. Pure, high quality and complete HMW-DNA suiting for construction of BAC library was obtained from the above leaf tissues.
Previous studies have shown that a clonal coverage of 6.0–8.0 genome equivalents is sufficient for coressponding research on genome-wide genetic dissection of complex organisms [20, 21, 23, 24]. Here, we have successfully constructed a G. mustelinum BAC library, which is large-insert, deep-coverage and lower empty vector rate. The high coverage BAC library contains all or most of the genomic information, which can be used for germplasm resources preservation permanently in view of the scarcity of G. mustelinum plant material [12].
In this study, the SSR marker Gh216 was used to screen the BAC library for specific BAC clones. This marker has once been used to obtain a subgenome-specific BAC clone enriched in repetitive sequences from the BAC library of G. herbaceum var. africanum [33]. The BAC clones identified by this marker may be candidate clones containing specific repetitive sequences. Here, using the bacteria liquid-PCR screening strategies, we identified 3 positive BAC clones from 96 super-pools containing 36,864 BAC clones, which indicate a higher positive clone screening rate from our BAC library.
At the same time, based on the constructed BAC library, studies on genome structure of G. mustelinum, including BAC-FISH-based karyotyping, physical mapping and even whole genomic sequencing, will be carried out, and all these works will serve as an important genomics resource to clone other genes conferring agronomically important traits efficiently.
Acknowledgments
This work was sponsored by the National Natural Science Foundation of China (31471548), National Key Research and Development Program (2016YFD0100203), Innovation Scientists and Technicians Troop Construction Projects of Henan Province (184200510009), Science and Technology Development Program of Henan Province (182102110048), Key Scientific Research Project of Higher Education in Henan Province (18B210001).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was sponsored by a grant from the National Natural Science Foundation of China (No. 31471548)(Renhai Peng), State Key Laboratory of Cotton Biology Open Fund (No. CB2017A06)(Yuling Liu), National Key Research and Development Program (2016YFD0100203)(Renhai Peng), PhD Scientific Research Fund of Anyang Institute of Technology (BSJ2016005)(Yuling Liu), Key Scientific Research Project of Higher Education in Henan Province (18B210001)(Yuling Liu), Science and Technology Development Program of Henan Province (182102110048), and Innovation Scientists and Technicians Troop Construction Projects of Henan Province (184200510009) (Renhai Peng). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fryxell PA. A revised taxonomic interpretation of Gossypium L., (Malvacea). Rheedea. 1992; 2: 108–165. [Google Scholar]
- 2.Wendel JF, Cronn RC. Polyploidy and the evolutionary history of cotton. Adv. Agron. 2003; 78: 139–186. doi: 10.1016/S0065-2113(02)78004-8 [Google Scholar]
- 3.Wendel JF, Grover CE. Taxonomy and evolution of the cotton genus, Gossypium Cotton, eds Fang D. D.and Percy R. G. (Madison, WI: American Society of Agronomy Inc.), 2015. pp. 25–44. doi: 10.2134/agronmonogr57.2013.0020 [Google Scholar]
- 4.Wendel JF, Rowley R, Stewart JM. Genetic diversity in and phylogenetic relationships of the Brazilian endemic cotton, Gossypium mustelinum (Malvaceae). Plant Syst Evol. 1994; 192: 49–59. https://doi.org/10.1007/BF00985907. [Google Scholar]
- 5.Grover CE, Zhu X, Grupp KK, Jareczek JJ, Gallagher JP, Szadkowski E, et al. Molecular confirmation of species status for the allopolyploid cotton species, Gossypium ekmanianum Wittmack. Genet Resour Crop Evol. 2015; 62: 103–114. doi: 10.1007/s10722-014-0138-x [Google Scholar]
- 6.Wang KB. Introduction and conservation of wild cotton in China. Cotton Sci. 2007; 19(5): 354–361. http://www.docin.com/p-1420587787.html. [Google Scholar]
- 7.Grover CE, Gallagher JP, Jareczek JJ, Page JT, Udall JA, Michael AG, et al. Re-evaluating the phylogeny of allopolyploid Gossypium L. Mol Phylogenet Evol. 2015; 92: 45–52. doi: 10.1016/j.ympev.2015.05.023 [DOI] [PubMed] [Google Scholar]
- 8.Wendel JF, Brubaker CL, Percival AE. Genetic diversity in Gossypium hirsutum and the origin of Upland cotton. Am J Bot. 1992; 79(11): 1291–1310. doi: 10.2307/2445058 [Google Scholar]
- 9.Wang BH, Wang W, Zhuang ZM, Zhu XY. Selection and evaluation of three Gossypium mustelinum near-isogenic lines. Chinese Agr Sci Bull. 2011; 27(24): 45–49. http://en.cnki.com.cn/Article_en/CJFDTotal-ZNTB201124009.htm. [Google Scholar]
- 10.Wang BH, Liu LM, Zhang D, Zhuang ZM, Guo H, Qiao X, et al. A genetic map between Gossypium hirsutum and the Brazilian endemic G. mustelinum and its application to QTL mapping. G3: Genes Genom Genet. 2016; 6: 1673–1685. doi: 10.1534/g3.116.029116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Q, Liu F, Li SH, Song GL, Wang CY, Zhang XD, et al. Uniqueness of the G. mustelinum genome revealed by GISH and 45S rDNA FISH. J Integr Plant Biol. 2013; 55(7): 654–662. doi: 10.1111/jipb.12084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang KB, Liu F. Research note on wild cotton in Brazil. China Cotton. 2014; 41(3): 6–7. [Google Scholar]
- 13.Ling P, Chen XM. Construction of a hexaploid wheat (Triticum aestivum L.) bacterial artificial chromosome library for cloning genes for stripe rust resistance. Genome. 2005; 48: 1028–1036. doi: 10.1139/g05-078 [DOI] [PubMed] [Google Scholar]
- 14.Schulte D, Ariyadasa R, Shi B, Fleury D, Saski C, Atkins M, et al. BAC library resources for map-based cloning and physical map construction in barley (Hordeum vulgare L.). BMC Genomics. 2011; 12: 247 doi: 10.1186/1471-2164-12-247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Liu Z, Peng R, Wang YH, Zhou ZL, Cai XY, et al. Cytogenetic maps of homoeologous chromosomes Ah01 and Dh01 and their integration with the genome assembly in Gossypium hirsutum. Comp Cytogene. 2017; 11(2): 405–420. doi: 10.3897/CompCytogen.v11i2.12824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohra A, Dubey A, Saxena RK, Penmetsa RV, Poornima KN, Kumar N, et al. Analysis of BAC-end sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment in pigeonpea (Cajanus spp.). BMC Plant Biol. 2011; 11:56 doi: 10.1186/1471-2229-11-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hulse-Kemp AM, Ashrafi H, Stoffel K, Zheng X, Saski CA, Scheffler BE, et al. BAC-end sequence-based SNP mining in allotetraploid cotton (Gossypium) utilizing resequencing data, phylogenetic inferences, and perspectives for genetic mapping. G3: Genes Genom Genet. 2015; 5(6): 1095–1105. doi: 10.1534/g3.115.017749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beier S, Himmelbach A, Schmutzer T, Felder M, Taudien S, Mayer KFX, et al. Multiplex sequencing of bacterial artificial chromosomes for assembling complex plant genomes. Plant Biotechnol J. 2016; 14(7): 1511–1522. doi: 10.1111/pbi.12511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong JM, Kohel RJ, Zhang HB, Yu J. Bacterial artificial chromosome (BAC) libraries constructed from the genetic standard of upland cottons. 1999. http://algodon.tamu.edu/htdocs-cotton/tm1bac.html. [Google Scholar]
- 20.Hu Y, Guo WZ, Zhang TZ. Construction of a bacterial artificial chromosome library of TM-1, a standard line for genetics and genomics in upland cotton. J Integr Plant Biol. 2009; 51(1): 107–112. doi: 10.1111/j.1744-7909.2008.00773.x [DOI] [PubMed] [Google Scholar]
- 21.Wang XF, Jun MA, Ma ZY, Zhang GY, Zheng YM. BAC library construction and characterization of Suyuan7235, a cotton germplasm with high fiber strength. Cotton Science. 2006; 18: 200–203. [Google Scholar]
- 22.Zheng YM, Wang XF, Zhang GY, Li XH, Ma ZY. BAC library construction of Zhongmiansuo12 with highyield, highquality and disease resistance. Journal of Agricultural University of Hebei. 2004; 27(3): 17–20. [Google Scholar]
- 23.Yin JM, Guo WZ, Zhang TZ. Construction and identification of bacterial artificial chromosome library for 0-613-2R in upland cotton. J Integr Plant Biol 2006; 48(2): 219–222. https://doi.org/10.1111/j.1744-7909.2006.00219.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang XF, Ma J, Wang WS. Zheng YM, Zhang GY, Liu CJ, et al. Construction and characterization of the first bacterial artificial chromosome library for the cotton species Gossypium barbadense L. Genome. 2006; 49(11): 1393–1398. doi: 10.1139/g06-113 [DOI] [PubMed] [Google Scholar]
- 25.Liu F, Wang YH, Gao HY, Wang CY, Zhou ZL, Cai XY, et al. Construction and characterization of a bacterial artificial chromosome library for the allotetraploid Gossypium tomentosum. Genet Mol Res. 2015; 14 (4): 16975–16980. doi: 10.4238/2015.December.15.3 [DOI] [PubMed] [Google Scholar]
- 26.Hu Y, Lu YM, Ma D, Guo WZ, Zhang TZ. Construction and characterization of a bacterial artificial chromosome library for the A-Genome of cotton (G. arboreum L.). J Biomed Biotechnol. 2011; 307–313. doi: 10.1155/2011/457137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao HY, Wang XF, Liu F, Peng RH, Zhang Y, Cheng H, et al. Construction of a bacterial artificial chromosome library for Gossypium herbaceum var. africanum. Chinese Sci Bull. 2013; 58(26): 3199–3201. doi: 10.1007/s11434-013-5864-5 [Google Scholar]
- 28.Wu YL, Wang YH, Liu YL, Wang CY, Cai XY, Wang XX, et al. Construction of a bacterial artificial chromosome (BAC) library from Gossypium raimondii. Mol Plant Breeding. 2016; 14(8): 2172–2177. doi: 10.13271/j.mpb.014.002172 [Google Scholar]
- 29.Cui XL, Liu F, Liu YL, Zhao YY, Zhou ZL, Zhao YY, et al. Construction of cytogenetic map of Gossypium herbaceum chromosome 1 and its integration with genetic maps. Mol Cytogenet. 2015; 8:2 doi: 10.1186/s13039-015-0106-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, Guan B, Guo WZ, Zhou BL, Hu Y, Zhu YC, et al. Completely distinguishing individual A-genome chromosomes and their karyotyping analysis by multiple bacterial artificial chromosome-fluorescence in situ hybridization. Genetics. 2008; 178: 1117–1122. doi: 10.1534/genetics.107.083576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gan YM, Liu F, Peng RH, Li SH, Zhang XD, Wang YH, et al. Individual chromosome identification, chromosomal collinearity and genetic-physical integrated map in Gossypium darwinii and four D genome cotton species revealed by BAC-FISH. Genes Genet Syst. 2012; 87: 233–241. doi: 10.1266/ggs.87.233 [DOI] [PubMed] [Google Scholar]
- 32.Wang K, Guo WZ, Yang ZJ, Hu Y, Zhang WP, Zhou BL, et al. Structure and size variations between 12A and 12D homoeologous chromosomes based on high-resolution cytogenetic map in allotetraploid cotton. Chromosoma. 2010; 119: 255–266. doi: 10.1007/s00412-009-0254-0 [DOI] [PubMed] [Google Scholar]
- 33.Liu YL, Peng RH, Liu F, Wang XX, Cui XL, Zhou ZL, et al. A Gossypium BAC clone contains key repeat components distinguishing sub-genome of allotetraploidy cottons. Mol Cytogenet. 2016; 9: 27 doi: 10.1186/s13039-016-0235-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang HB, Zhao XP, Ding XL, Paterson AH, Wing RA. Preparation of megabase-size DNA from plant nuclei. Plant J. 1995; 7: 175–184. doi: 10.1046/j.1365-313X.1995.07010175.x [Google Scholar]
- 35.Hendrix B, Stewart JM. Estimation of the nuclear DNA content of Gossypium species. Ann Bot. 2005; 95: 789–797. doi: 10.1093/aob/mci078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang LK, You WW, Zhang XJ, Xu J, Jiang YL, Wang K, et al. Construction of the BAC library of small abalone (Haliotis diversicolor) for gene screening and genome characterization. Mar Biotechnol 2016; 8(1): 49–56. doi: 10.1007/s10126-015-9666-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.