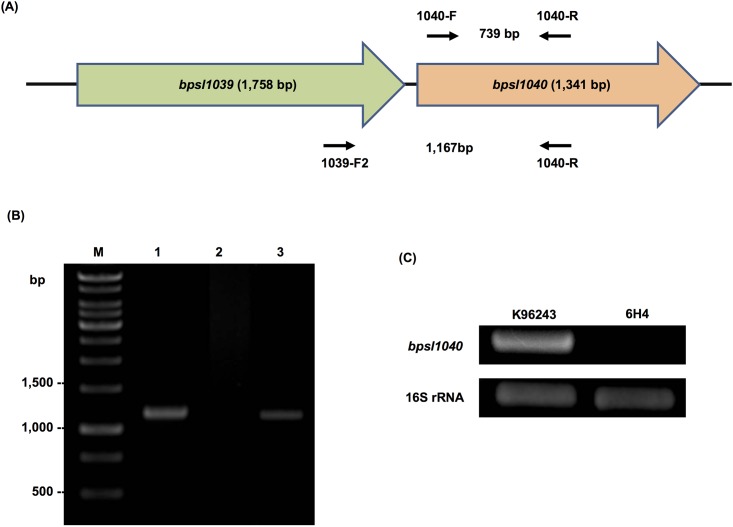
Fig 1. Gene co-localization of B. pseudomallei bpsl1039-bpsl1040 and RT-PCR analysis.
(A) Organization of B. pseudomallei bpsl1039-bpsl1040 genes and the location of primer pairs used in RT-PCR analysis. (B) RT-PCR analysis using primers 1039-F2 and 1040-R showed co-transcription of B. pseudomallei bpsl1039-bpsl1040 genes (lane 3). Lanes 1 and 2 represent positive and negative controls, respectively, using wild-type genomic DNA and the extracted RNA, respectively. A negative RT-PCR control (lane 2) confirms that the band observed in the positive reaction is not DNA contamination. (C) RT-PCR analysis of bpsl1040 expression, using primers 1040-F and 1040-R, was performed in B. pseudomallei wild-type (K96243) and 6H4 mutant. The 6H4 mutant showed the absence of bpsl1040 expression. B. pseudomallei 16S rRNA gene was amplified as control. Lane M represents 1 Kb DNA marker ladder.