SUMMARY
Voltage-gated ion channels (VGICs) are associated with hundreds of human diseases. To date, 3D structural models of human VGICs have not been reported. We developed a 3D structural integrity metric to rank the accuracy of all VGIC structures deposited in the PDB. The metric revealed inaccuracies in structural models built from recent single-particle, non-crystalline cryo-electron microscopy maps and enabled the building of highly accurate homology models of human Cav channel α1 subunits at atomic resolution. Human Cav Mendelian mutations mostly located to segments involved in the mechanism of voltage sensing and gating within the 3D structure, with multiple mutations targeting equivalent 3D structural locations despite eliciting distinct clinical phenotypes. The models also revealed that the architecture of the ion selectivity filter is highly conserved from bacteria to humans and between sodium and calcium VGICs.
In Brief
Martinez-Ortiz et al. develop an accuracy metric for structural models of voltage-gated ion channels based on the conservation of a core functional cluster of amino acids. The significance of the method is its ability to reveal and correct subtle modeling errors and refine low-resolution VGICs to atomic accuracy.
INTRODUCTION
The pore-forming α1 subunit of voltage-gated ion channels (VGICs) is composed of four repeats (I, II, III, and IV), each containing six membrane-spanning α helices often referred to as segments (S1–S6). In 3D space, the S1–S4 cluster into an intra-repeat voltage-sensing domain (VSD), and S5 and S6 from all four repeats interlock to form an inter-repeat pore domain (PD) through which the ion is transmitted. A cluster of positive residues in S4 is known as the “voltage sensor” (VS). (Yu and Catterall, 2004) Mutations in these segments within the α1 subunit of Cav channels are associated with multiple heritable human diseases, including episodic ataxia type 2, familial hemiplegic migraine type 1, congenital stationary night blindness type 2, and other autosomal dominant diseases (Adams and Snutch, 2007; Bech-Hansen et al., 1998; Caciotti et al., 2003; Chang et al., 2006; Chen et al., 2003; Denier et al., 1999; Fouad et al., 1997; Heron et al., 2004; Jen et al., 1999; Jeng et al., 2006; Jurkat-Rott et al., 1994, 2002; Khosravani et al., 2005; Mantuano et al., 2004; McRory et al., 2004; Ophoff et al., 1996; Ptácek et al., 1994; Spacey et al., 2004; Splawski et al., 2004, 2005, 2006; Striessnig et al., 2004; Strom et al., 1998; Vitko et al., 2005; Wallace, 1986; Wappl et al., 2002). An enormous number of additional phenotypes across many species also map to VGIC α1 subunits. To better understand the mechanisms of each human channel and biophysical structure-activity relationships (SARs) associated with inherited (Mendelian) disease-causing mutations, an atomic-detailed 3D view of the relative arrangement of amino acids in the pore-forming repeats and segments of the α1 subunits of the human Cav family is necessary (Van Petegem and Minor, 2006).
We applied the bioinformatics concept of “mutual information” (Dunn et al., 2008; Kamisetty et al., 2013; Tress and Valencia, 2010) to construct a metric capable of assessing the structural accuracy of all VGIC X-ray crystal and cryo-electron microscopy (cryo-EM) structures deposited in the PDB. Because the metric is a quantitative score that can be applied to any VGIC model, it can be used to guide and assess the value of individual molecular modeling steps (target-template sequence alignment, side chain building, loop building, etc.) and, therefore, build maximally accurate 3D models of the α1 subunit for any VGIC. We employed our metric to build structural models of human Cav channel α1 subunits. Because the best available X-ray crystallographic VGIC structures are resolved at atomic resolution (<3.5 Å), our models, which inherit conserved architectural features from these structures, are resolved at that same atomic level of detail and may be of improved utility for SAR studies. Accordingly, we used our models to visualize the 3D structural locations of disease-causing mutations that target this family of channels and to evaluate the extent of architectural conservation in the selectivity filter among the family of human Cav channels and homologous VGICs.
RESULTS
The Contact Area of Conserved VSD Side Chains Reflects VGIC Structural Integrity
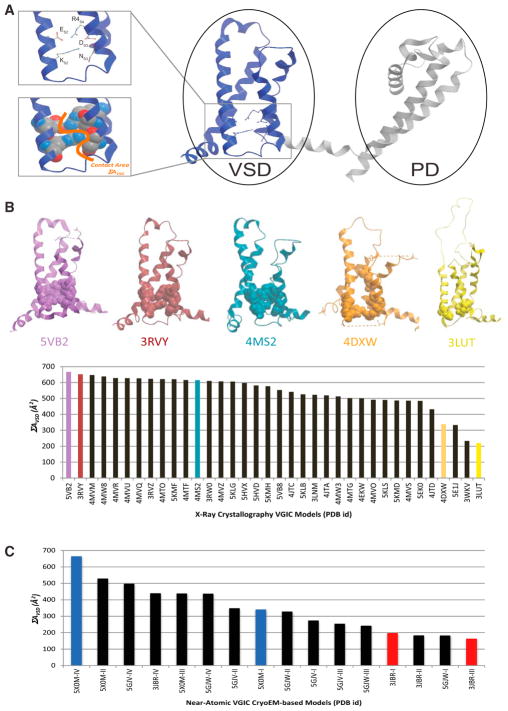
A tripartite, discontinuous, tertiary intra-molecular interaction between S2, S3, and the VS (S4) is conserved and structurally evident within the hydrophobic core of the VSD of all VGIC structures reported in the PDB (Figure 1; Payandeh et al., 2011). This interaction forms a network of salt bridges between VGIC-conserved FxxExxxR and SxxD motifs of S2 and S3, respectively, and the last positive charge of the VS (S4), consisting of the following contacts: S2 forms one intra-segment salt bridge with itself (ES2-RS2), one inter-segment salt bridge with S4 (ES2-R4-S4), and two inter-segment salt bridges with S3 (RS2-DS3 and KS2-SS3), whereas S3 forms a third inter-segment salt bridge with S4 (DS3-R4-S4), as depicted in Figure 1A. The amino acids that participate in this network or structural motif are conserved in all VGIC VSD domain sequences and structures reported, despite significant variations in amino acid identity at directly adjacent positions (Figure S1; Guda et al., 2007; Yu and Catterall, 2004). Accordingly, we defined the total surface contact area between this ensemble of interacting side chains (ΣAVSD) as an accuracy metric for any VSD 3D structural model in any VGIC.
Figure 1. ΣAVSD Value Scores the Preservation of Evolutionarily Conserved Interactions of the VSD in All VGIC Structures Deposited in the PDB.
(A) All voltage-gated ion channels (VGICs) contain a conserved structural motif within the voltage-sensing domain (VSD) as depicted by the ribbon, stick, and CPK representation of the VSD structural motif of the sodium NavAb voltage-gated channel (PDB: 3RVY), where a unique cluster of side chains forms a tripartite salt bridge network that connects amino acids from S2, S3, and S4, creating a clear ΣAVSD signature (orange line) for the VSD.
(B and C) Contact area values (ΣAVSD) of VGIC X-ray crystal structures (B) and cryo-EM-based models (C), showing the conservation of the VSD motif in all structures deposited with the more resolved structures achieving higher ΣAVSD values. The structures are identified in the graphs by their unique PDB identifying codes.
See also Figure S1.
ΣAVSD Ranking of All VGIC Structures Deposited in the PDB
Searching the PDB with the keyword “voltage-gated channel” yielded over 200 X-ray crystallographic structural models, fewer than 40 of which encompassed the full VSD. Defining the ΣAVSD for all experimentally visualized VSDs revealed a correlation between high ΣAVSD values and structures with higher overall percentile scores of global validation metrics of structural quality (Figure 1B; Table 1). A plateau appears at ~500–600 Å2 ΣAVSD, suggesting that this is an accuracy threshold for the experimental resolution of VSD domains in VGICs. ΣAVSD ranges from 320–600 Å2 among structures with the intact cluster (acceptable range), suggesting that ΣAVSD captures different conformations of the VS; namely, up and down conformations. Our statistical analyses show that an ΣAVSD value of 320 Å2 discriminates between acceptable and unacceptable structures (p = 1.14 × 10−17; Experimental Procedures), with the ΣAVSD score greatly degrading for inaccurate/less resolved X-ray crystal structures such as PDB: 4DXW, PDB: 5E1J, and PDB: 3LUT, where the key cluster of residues is disrupted. The same analysis performed on VGICs built from single-particle cryo-EM maps deposited in the PDB revealed much poorer ΣAVSD scores that did not correlate with global validation metrics of structural quality applicable to cryo-EM models (Table 2). Specifically, recent models of rabbit Cav1.1 α1 subunits built from single-particle cryo-EM maps (PDB: 3JBR, Wu et al., 2015; PDB: 5GJV, Wu et al., 2016) exhibited unacceptable ΣAVSD scores (≤320 Å2 ΣAVSD), whereas a model of Periplaneta americana NavPaS (PDB: 5X0M) (Shen et al., 2017), a voltage-gated sodium channel (VGSD) solved by the same authors, reported lower general structural quality validation metric scores but exhibited an acceptable ΣAVSD score (Table 2; Figure 1C).
Table 1.
Global Accuracy Metric Percentile Scores of VGIC VSD X-Ray Structures as Provided by the PDB
| PDB Code | Resolution (Å) | R-Free | Clash Score | Ramachandran Outliers | Side-Chain Outliers | RSRZ Outliers |
|---|---|---|---|---|---|---|
| 3LUT | 2.9 | 0.222 | 23 | 2.50% | 6.80% | 20.30% |
| 3RVY | 2.7 | 0.289 | 32 | 0.20% | 4.20% | 5% |
| 4DXW | 3.05 | 0.261 | 35 | 5.40% | 16.10% | 1.60% |
| 4MS2 | 2.75 | 0.25 | 16 | 0.50% | 8.20% | 2.90% |
| 5VB2 | 3.2 | 0.27 | 6 | 0.00% | 3.10% | 2.10% |
This table contains the global percentile scores of the data and model accuracy of VGIC crystal structures containing the VSD, as listed in the summary report of the database. The data quality validation metrics include free R-factor (R-Free) and Clash Score and Ramachandran, side-chain, and real-space R-value Z-score (RSRZ) outliers as reported for each selected example structure in the Structure Summary section of the database. See also Figures 1 and 2.
Table 2.
Global Accuracy Metric Percentile Scores of VGIC VSD Single-Particle Cryo-EM Structures as Provided by the PDB
| PDB Code | Resolution (Å) | Clash Score | Ramachandran Outliers | Side-Chain Outliers |
|---|---|---|---|---|
| 3JBR | 4.2 | 19 | 7.10% | 0.50% |
| 5GJV | 3.6 | 18 | 2.00% | 4.80% |
| 5X0M | 3.8 | 53 | 5.20% | 12.20% |
Listed in this table are the global percentile scores of the data and model accuracy of VGIC cryo-EM-based models containing the VSD, as listed in the summary report of the PDB. The data quality validation metrics include Clash Score and Ramachandran and side-chain outliers as reported for each selected example structure in the Structure Summary section of the database. See also Figure 2.
ΣAVSD Scores Correlate with Inaccuracies in Residue Assignment and Provide Structural Constraints for Accurate Homology Modeling of VGIC VSDs
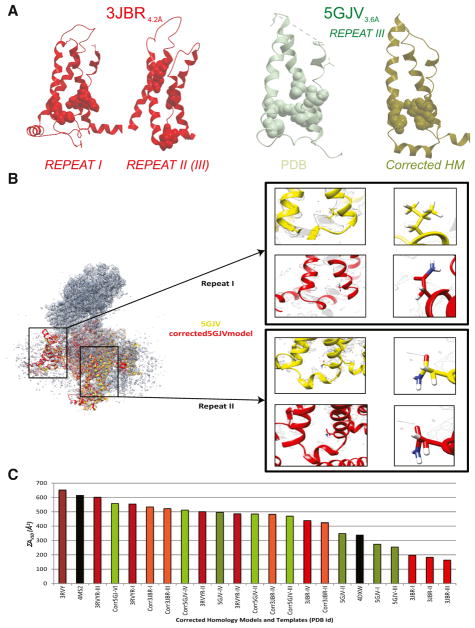
Analysis of the residue assignments in the two recent cryo-EM-derived Cav1.1 α1 subunit 3D structural models exhibiting poor ΣAVSD scores revealed unambiguous errors in the placement of VSD motif side chains within the cryo-EM-based models (Figure 2A). The specific list of inaccurate residue assignments is shown in Table 3. Inaccurate residue assignment of the cryo-EM-based models appears to be a direct result of inaccurate model building in sparser areas of the experimental map. These inaccuracies in residue assignments result in low ΣAVSD scores for these specific domains/repeats because the tripartite interaction is impossible with the alternate location of the amino acids of the VSD motif. In addition, because the ΣAVSD scores improved dramatically for the accurate repeats of PDB: 5GJV (repeats II and IV) versus its incorrect repeats (PDB: 5GJV, repeats I and III) and incorrect repeats in the identical protein from a lower (PDB: 3JBR) resolution dataset, ΣAVSD appears to be able to distinguish errors and inaccuracies in VGIC models, including those deriving from avoidable modeling errors, which are not evident with any other metrics, such as root-mean-square deviation (RMSD; Figure S2). Although ΣAVSD only pertains to the VSD, accuracy in modeling this domain is highly correlated with structural accuracy of the entire subunit, including the PD, because the sequence signal of the PD is so high that unambiguous sequence alignment and modeling of this domain are relatively trivial tasks for most of the template structures available.
Figure 2. ΣAVSD Scores Correlate with Inaccuracies in Residue Assignment and Provide Structural Constraints for Accurate Homology Modeling of VGIC VSD.
(A–C) Ribbon, stick, and CPK representation of the VSD structural motif of PDB: 3JBR and PDB: 5GJV (A), both near-atomic cryo-EM structures of the Cav1.1 channel that lack the VSD interaction as a result of incorrect residue assignment of the functional residues. For PDB: 5GJV, we show that, by correctly placing the functional residues in their accurate locations, we reconstitute the VSD interaction in our corrected structure and produce a molecular model that is in higher agreement with the cryo-EM electron density map and raw data, as depicted (B) by the structural superimposition of the deposited 3D cryo-EM-based model (PDB: 5GJV) and our corrected-5GJV homology model, which shows that our model’s residue assignment correlates better with the cryo-EM density map than the deposited model. This is evidenced by the specific density pattern created by glutamate versus that of leucine, in which the oxygen atoms are expected to be distorted whereas the carbon atoms are expected to be resolved in the electron density map, as highlighted by the enlargements of the same amino acids in repeat I, which was incorrectly modeled, and repeat II, which was correctly modeled. Our corrected rabbit Cav1.1 model not only correlates better with the map, but also displays an improved ΣAVSD score over the models that have been deposited in the PDB, as shown in the graph (C). See also Figure S2.
Table 3.
Modeling Errors in Recent VGIC VSD Cryo-EM-Based Models
| PDB Code | Resolution (Å) | Repeat I | Repeat II | Repeat III | Repeat IV |
|---|---|---|---|---|---|
| 3JBR | 4.2 | C-term translation: S3 (2 pos.) | N-term translation | N-term translation | errors in side-chain angles (16° difference) |
| S2 (7 pos.); | S2 (7 pos.); | ||||
| S3 (5 pos.) | S3 (5 pos.) | ||||
| 5GJV | 3.6 | C-term translation: S3 (5 pos.) | no errors | C-term translation: S3 (3 pos.) | no errors |
| 5X0M | 3.8 | no errors | no errors | no errors | no errors |
Listed in this table are the direction and magnitude of frameshift molecular modeling errors of recent cryo-EM-based models of VGIC; PDB: 3JBR, 5GJV, and 5X0M. For each repeat in the tetrameric subunit, the inaccuracies in residue assignment are described for S2 and S3 of the VSD in the structural models. This table also lists the resolution of the cryo-EM map to emphasize that the modeling errors are exclusive to low-quality maps, as determined by resolution. C-term, C terminus; N-term, N terminus; pos., positions. See also Figure 2.
Interestingly, models of the rabbit Cav1.1 α1 subunit that are significantly more accurate than the 3.6-Å PDB: 5GJV model could be generated directly from the 4.2-Å PDB: 3JBR map using the ΣAVSD score. ΣAVSD easily identifies the best template for homology modeling, and the unorthodox four-repeat structure can easily be built by faithfully applying sound theoretical principles of homology modeling, using the VSD motif as a structural landmark for molecular modeling restraints (as described in the Experimental Procedures). Accordingly, we used our method to correct the remaining modeling errors in the best rabbit Cav1.1 α1 subunit cryo-EM molecular model (PDB: 5GJV) and produce a 3D model that more accurately conveys the density acquired in the cryo-EM map for the functional side chains. We observed that our “corrected model” of PDB:5GJV (Figure 2B) achieved an improved ΣAVSD score, equivalent to those characteristic of the highest (atomic)-resolution VGIC structures (Figure 2C). We then used this maximally accurate, corrected rabbit Cav model of the α1 subunit as a structural template to construct homology models of the α1 subunits of the entire human Cav family, which are themselves at the same maximal (atomic) resolution and level of accuracy (Figure S3). The ΣAVSD score and, independently, the sequence similarity between the rabbit Cav1.1 template and the human Cav α1 subunits are sufficiently high, that the human Cav models we have built may be considered the highest atomic-resolution structural models currently available (Experimental Procedures; Abagyan and Batalov, 1997; Abagyan and Totrov, 1997; Cardozo et al., 1995).
Mapping of Genetic Mutations Associated with Clinical Phenotypes onto 3D Models of Human Cav Channels
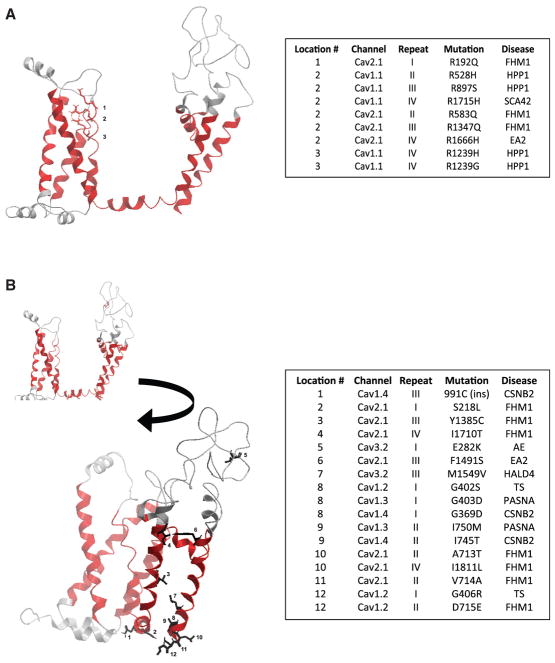
Human Cav channels are associated with a large number of missense Mendelian mutations that have distinct clinical phenotypes. The symmetry of the repeats in these proteins allowed us to structurally equate specific amino acid positions of the α1 subunit in different repeats and different family members. This approach enabled the mapping of missense Mendelian mutations into a single universal structural context independent of the specific channel, repeat, or disease associated with the mutation, as depicted in Figure 3. Our approach and models reveal that phenotypically distinct diseases target equivalent 3D structural sites within the highly conserved structural regions of the channels, which have been previously associated with the mechanistic properties of voltage sensing and gating.
Figure 3. Mapping of Genetic Mutations Associated with Clinical Phenotypes onto 3D Models of Human Cav Channels.
(A and B) Mapping of a representative group of disease-causing mutation onto the 3D model of human Cav1.1 repeat I, as depicted with the ribbon and stick representation of the structure. The legend table shows a large number of mutations targeting both the voltage-sensing (A) and gating (B) mechanisms of the channels and that multiple mutations target the same structural location while leading to an array of phenotypically diverse diseases. The pseudo-numbering of the mutations only takes into account the mutated sites within the domains and begins at the first mutated amino acid. See also Figure S3 and Table S1.
Mutations Affecting Voltage Sensing
Many pathogenic mutations that affect the voltage-sensing mechanism of the channels were located in the segments of the VSD (S1–S4). While some mutations located in other segments have also been associated with aberrant voltage sensing. We subdivided the mutations targeting the VSD into two categories for the purposes of discussion: mutations targeting the VS (S4) segment itself (Figure 3A) and mutations mapping to the other segments (S1–S3) of the VSD.
Four representative autosomal diseases—familial hypokalemic periodic paralysis (FHPP1, Cav1.1), episodic ataxia type 2 (EA2, Cav2.1), familial hemiplegic migraine type 1 (FHM1, Cav2.1), and spinal cerebellar ataxia 42 (SCA42, Cav3.1)—exhibited mutations mapping to the VS (S4) segment of three different channels (Figure 2). In general, the mutations that map to this segment: target the positively charged residues, reduce the number of positive residues (from a positively charged amino acid to a shorter, non-positively charged amino acid), and preferentially mutate the second positive residue (R2) of the VS. The only exceptions are R1664H in Cav2.1 associated with episodic ataxia 2 and R195K in Cav1.2 associated with FHM, which significantly modify the structure of the chemical group while preserving the positive charge of the terminal side chains (Figure 3).
Mutations mapping to segments S1–S3 of the VSD are difficult to interpret given the limited experimental data linking the specific sites of mutated amino acids to specific mechanistic functions of the channels. However, three inherited diseases are caused by mutations targeting these segments: FHPP1 (Cav1.1), absence epilepsy (AE, Cav3.2), and epileptic encephalopathy early infantile 42 (EIEE42, Cav3.2). Interestingly, among these, two similarly located or “functionally equivalent” mutations, Cav1.1_III V876E and Cav3.2_III V831, lead to two phenotypically distinct diseases: FHPP1 and AE (Table S1).
Mutations Affecting Gating
Mutations in the S4-S5 linker and S5 and S6 segments are referred to as mutations that affect the gating mechanism of the channels. The S4-S5 linker, although not a segment but an inter-segment linker, is highly conserved among the family, mechanistically proven to be part of the gating assembly of the channels, and embedded in the membrane, as suggested by recently published structures. Seven representative diseases exhibited mutations targeting the S5 and S6 segments and have been associated with affecting the gating mechanism of the channels: Timothy syndrome (TS, Cav1.2); primary aldosteronism, seizures, and neurological abnormalities (PASNA, Cav1.3); congenital stationary blindness type 2 (CSNB2, Cav1.4); FHM1 (Cav2.1); EA2 (Cav2.1); AE (Cav3.2); and hyperal-dosteronism type 4 (HALD4, Cav3.2) (Figure 3B). The mutated sites typically targeted helical points of contact within the gate-forming segments and introduced electrostatic side chains within the hydrophobic areas of the helical edges. Mutations in these segments were found in all voltage-gated calcium channel (VGCC)-exhibiting clinical phenotypes, with the exception of Cav1.1 and Cav3.1. Most mutations were located at the tip of S6. Interestingly, the sites mutated in TS are repeatedly targeted by mutations causing other diseases. Here we note 3 distinct diseases (PASNA, CSNB2, and FHM1) with mutations mapping to this same structural location, representing a SAR between 4 different channels and phenotypes.
Architecture of the Selectivity Filter in Human Cav Channels
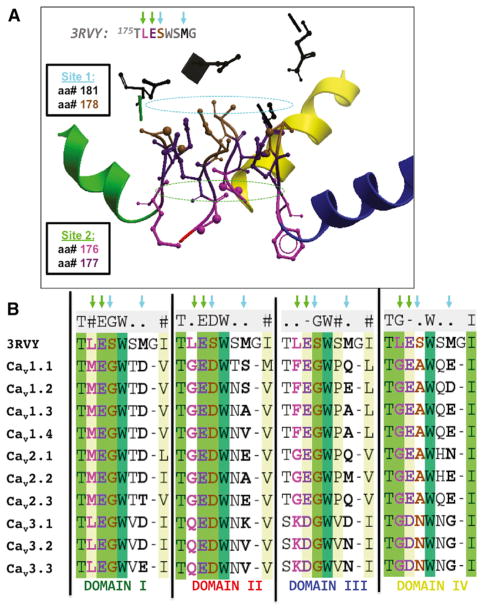
In addition to mapping disease-causing mutations, the accuracy of our models allows us to assess the extent of architectural conservation over the vast evolutionary distance between mammalian calcium (rabbit and human Cav) and bacterial sodium (NavAb) selectivity filters of homologous ion channels (Payandeh et al., 2011, 2012; Tsai et al., 2013; Zhang et al., 2012). The mechanism of Ca2+ permeation and the architecture of the ion selectivity filter, which was revealed by the crystal structure of CavAb and named, from extracellular to intracellular, sites 1, 2, and 3 is largely conserved in mammalian Cav channels (Figure 4; Catterall et al., 2017; Tang et al., 2014).
Figure 4. The Architecture of the Selectivity Filter Is Conserved in Human Cav Channels.
(A) Structural side view of 3 repeats in the human model of Cav1.1 shows the conservation in space of the amino acids that compose the 3 selectivity rings initially described for the CavAb-engineered channel, as depicted by the ribbon and stick representation of the subunits.
(B) In the multiple sequence alignment, we observe that the residues involved in the selectivity filter sites: 1, 2, and 3, initially described for CavAb, are conserved in all human Cav channels with varying amino acids participating in each site.
See also Figure S3.
Specifically, the atoms seen in CavAb contributing to sites 2 and 3 are conserved both in sequence and in 3D space in all of our modeled Cav channels; however, the 4-fold symmetric planar ring of site 1 appears to be formed distinctly across the different channels. In Cav1.x and Cav2.x, site 1 is formed with the terminal side chain of one amino acid located at an equivalent position in both sequence and structure to position 178 in CavAb and three terminal side-chain atoms originating from the amino acid position equivalent to 181 in CavAb. For Cav3.x, site 1 is constructed by two amino acids at the location equivalent to CavAb’s 178 amino acid and two amino acids from amino acid location equivalent to CavAb’s 181, as illustrated in Figure 4. The unusual exceptions are Cav1.3 and Cav1.4, where three, not four, charged amino acids are found mapping to site 1. Thus, the 3D architecture of the selectivity filter is largely conserved but is built from different contributing amino acid side chains. These findings suggest that contributing amino acids to this site have varied through evolution and that this variation itself differs from repeat to repeat across the family, as depicted in the aligned sequences of Figure 4.
DISCUSSION
We report the contact area (ΣAVSD) score for VGIC, a unique structure quality metric capable of scoring the accuracy of VSD residue assignment and, therefore, the accuracy of the whole α1 subunit for any VGIC structure regardless of the origin and quality of the experimental data. Our metric therefore standardizes diverse X-ray and cryo-EM model quality validation scores, with our results showing that the ΣAVSD score enables the refinement of a near-atomic VGIC single-particle, non-crystalline, cryo-EM dataset to atomic resolution (Figure 4C), which is a tremendous gain in value for any near-atomic cryo-EM data-set. Failure to utilize this ΣAVSD approach and adhere to sound theoretical homology modeling principles has the potential of wasting significant experimental signal. Notably, common automated homology modeling algorithms fail to produce correct models (Table S2). Thus, the use of ΣAVSD is proposed as a critical component of any experimental VGIC structure determination attempt, many of which are currently underway in the field.
The ΣAVSD metric has enabled the building of state-of-the-art homology models of the human α1 subunits of the Cav family of VGIC at the equivalent of the highest possible (atomic) resolution, which is arguably a superior resolution and accuracy to all but one recent single-particle, non-crystalline, cryo-EM-based VGIC α1 subunit structure. Because of their accuracy, these models have multiple applications. Here we used the models to better understand the mechanism of disease-causing heritable mutations that target the α1 subunit. We classified a representative group of the mutations according to their locations within state-of-the-art structural models of human Cav channels, which revealed that the segments associated with voltage sensing (S4), pore formation (P-loops), and gating (S4-S5 linker, S5, and S6) are preferentially mutated compared with other segments of the channel.
By superimposing all repeats of all human Cav channels, we noticed that multiple disease-causing mutations map to the same segments and even to equivalent residues between the human channels while leading to distinct disease phenotypes. Some examples are S229P in Cav1.4 and S218L in Cav2.1, which map to the same location within the structure but are associated with different diseases (S229P, CSB2; S218L, FHM1); G402S in Cav1.2 and G369D in Cav1.4, which lead to TS and CSNB, respectively; and lastly, R528H in Cav1.1, associated with FHPP, R195K, R583Q, and R1347Q in Cav2.1, associated with FHM; and R1666H, also in Cav2.1 but associated with EA2; all map to the same structural site: the first positive residue of the S4 segment.
We also noted that some diseases were associated with multiple mutations and exhibited a homogeneous distribution of mutations throughout the structure, with no functional elements preferentially mutated, such as CSNB, EA2, and FHM, whereas other diseases, such as TS, preferentially mutate a single mechanistic element: the gate. These differences would suggest that some VGCC diseases, such as those exhibiting a homogeneous distribution of the mutations, may be associated with reduced or increased stability of the channel or with specific post-translational effects on expression, whereas other diseases, such as TS, that exclusively target the gate would suggest a direct correlation between a specific functionality of the channel and the disease.
As expected, some mutations target the same structural location in different channels and lead to distinct diseases. Such mutations may lead to the same electrophysiological effect in both channels, but the observable disease phenotypes may ultimately depend, at least in part, on the cellular or tissue context (e.g., the differential expression of the channel in body and brain tissues) and on the effects of these mutations on the divergent, non-α1 subunits of the channels. Other interesting mutations were found to target the same channels at equivalent structural sites while leading to different diseases. The convergence of diseases with dramatically different phenotypes targeting the same functional element, or even to the same amino acid, in different repeats of the same channel suggests domain-specific functions for equivalent sites within the repeats. Thus, differential cellular or tissue contexts of channels may not be the only mechanism whereby different family members produce different phenotypes; structure-phenotype relationships also appear to be present and dependent on specific architectural features of the channel.
The location of disease-causing mutations within these homology models identifies relationships between otherwise distant diseases by grouping them from a novel 3D structural/biophysical point of view according to the functional elements affected. The confidence accuracy of our models, conferred by ΣAVSD, also allowed us to establish a high degree of architectural conservation of the selectivity filter between bacterial sodium VGICs and mammalian calcium VGICs, where three specific binding sites are conserved but constructed differentially for the different channels. The availability of the 3D models and the data presented here, which relate to the overall architecture of the channels and to disease-causing mutations, is thus expected to serve as a previously unavailable 3D structural reference map for many investigations into the mechanism and function of this family of channels in health and disease.
EXPERIMENTAL PROCEDURES
ΣAVSD Metric
The ΣAVSD between amino acid residues was calculated with ICM-Pro as described previously (Abagyan and Totrov, 1997). Briefly: for any two residues (i and j) in a full-atom, 3D structural model, a Connolley surface around the residue (i) is generated by rolling a suitably sized probe over the van der Waals surface of the residue atoms and tracing the surface encountered by the center of the probe. The angstrom-squared surface of residue (i) occluded by the analogous surface residue (j) is the ΣAVSD for that pair. The aggregate ΣAVSD for any arbitrary set of residues can be summed. In this case, only the amino acids participating in the salt bridges of the VSD motif were summed to generate ΣAVSD.
VGIC PDB Search
To identify all deposited structures that contain the VSD motif, we searched the PDB with the term “voltage-gated channel” and filtered the results to select only X-ray and cryo-EM structures. Multiple sequence alignments of the resulting hits (240 total) were generated using the ClustalW method, with 20–40 sequences at a time. After identifying the ΣAVSD motif in the multiple sequence alignment, the amino acids were located in the 3D structural model to ensure that they co-locate in the canonical 4-helix VSD bundle and are, in fact, all facing toward the hydrophobic core of the domain, forming a single intramolecular (intradomain) cluster. For the structural models that contained the structural motif, we recorded the residue numbers and calculated the corresponding ΣAVSD.
Statistical Methods
Because ΣAVSD consists of continuous data, two-tailed Student’s t test was calculated to assess the statistical significance of the difference in ΣAVSD score between the “acceptable” (>320-Å2 ΣAVSD) group of X-ray and cryo-EM structures and the “unacceptable” (<320-Å2 ΣAVSD) group.
Homology Modeling
Sequence Alignment
All sequence alignments were performed with a ClustalW-based algorithm in the ICM-Pro software package (Molsoft, La Jolla CA) (Cardozo et al., 1995).
Model Building and Refinement
Model building was performed as described previously using the following steps in ICM-Pro (Molsoft, La Jolla, CA) (Cardozo et al., 1995). Briefly, an extended polypeptide chain with idealized covalent geometry (Abagyan and Totrov, 1994) was built for each of the four repeats of the α1 subunit. Torsion angles in each amino acid in each Cav repeat were assigned the same numerical value as observed for their counterparts in the template PDB crystallographic structure, using pairwise sequence alignment as a guide to establish the amino acid equivalences between repeat and template. The most likely rotamer for each side chain in each amino acid was assigned to non-identical aligned residues, and near-extended backbone torsion values were assigned to the inserted or deleted (un-aligned) amino acid residues. The sum of the physical energy and quadratic restraints between corresponding atoms in the model and template structures were iteratively minimized, reducing the strength of the restraint potential with each iteration. Non-identical side chains and loops were subjected to biased probability Monte Carlo sampling (Abagyan and Totrov, 1994) to produce the lowest energy structure matching the template coordinates. For the human model, each Cav repeat was built on a successive clockwise arrangement around the pore, with the order of repeats established with analysis of cryo-EM density data.
3RVY-R Modeling
Structural Template Selection
The crystallographic structure of the voltage-gated Na+ channel from Arcobacter butzleri (NavAb) resolved to 2.7 Å (PDB: 3RVY) was used as the template to build an initial model of the rabbit Cav1.1 α1 subunit (referred to as the “3RVY-R model”)(Payandeh et al., 2011). We selected this template because it received the highest ΣAVSD score while also resolving an entire repeat (voltage-sensing and PDs) at atomic resolution.
Sequence Alignment
Pairwise zero-end-gap global sequence alignments (ZEGAs) resulted in a p value for confidence of structural similarity between rabbit Cav1.1 (CAC1S, UniProt: P07293) and 3RVY averaged p < 10−12.5 (Abagyan and Batalov, 1997), confirming that correctly executed homology modeling would likely yield a degree of structural accuracy equivalent to the NavAb template (2.7 Å).
Challenges in Modeling 3RVY-R
The first challenge was modeling the “heterotetrameric” subunit, formed by four internal non-identical VSD-PD repeats, with a homotetrameric template (NavAb, PDB: 3RVY) (Payandeh et al., 2011) where four identical VSD-PD chains form the subunit. We met this challenge with a multilink approach (Rost, 1999). First, we identified that rabbit Cav1.1 repeat II had the highest similarity to 3RVY. Thus, the domain boundaries of repeat II of Cav1.1 were clearly delineated, and an initial model of rabbit Cav1.1 repeat II was built based on 3RVY’s atomic coordinates. We noted that Cav1.1 repeats I, III, and IV have a higher similarity to repeat II than to the 3RVY template. Consequently, we used ZEGA global pairwise alignments (Abagyan and Batalov, 1997) between repeat II and the remainder of the Cav1.1 sequence to find and build models of the remaining three internal repeats of the rabbit Cav1.1 α1 subunit using the structural model of repeat II as a template.
A second challenge to modeling 3RVY-R “blinded” to the recent single-particle, non-crystalline cryo-EM data was determining the order of the repeats around the ion-conducting pore. The order of the repeats for our homology model was initially determined by analyzing the non-atomic EM density pattern reported for human Cav3.1 (Walsh et al., 2009). Briefly, from the Cav3.1 cryo-EM density pattern resolved at 20 Å, we noted that a C-terminal gold probe nestled around the N terminus of repeat I in a counter-clockwise manner as viewed from the intracellular surface, which suggests a clockwise orientation of the repeats as viewed from the extracellular surface, as reported by Walsh et al. (2009). Each Cav repeat in our 3RVY-R model was thus built on a successive clockwise arrangement around the pore. To achieve the modeling of the repeats around the pore, we independently modeled each unlinked repeat based on the 3RVY 3D coordinates and used the crystal contact points of the deposited structure, where a symmetric tetramer was generated by the crystal lattice, to superimpose and, hence, guide the modeling of each repeat to a clockwise orientation as viewed from the extra-cellular side of the model.
Residue Assignment Optimization of 5GJV
Residue Assignment/Structure Comparison
3D structural visualization and sequence alignment were used to compare the residue assignment of all rabbit Cav1.1 models. To identify the residues assigned to each of the segments (S1–S6, 24 segments in total in each model), we visually inspected all transmembrane helices from all structural models and noted the residue number for the first and last 3 amino acids of the selected helix. We then proceeded to draw a box in each protein sequence around the amino acids that corresponded to the contiguous stretch of amino acids between the first and last 3 amino acids of the helix observed.
Refinement of the Cryo-EM-Based PDB Structures Using the 3RVY-R Model
Analyzing the cryo-EM data, we noted that the order of the repeats of the subunit was corroborated by the report of the rabbit cryo-EM 4.2-Å structure (PDB: 3JBR) via the analysis of the cryo-EM densities created by the varying lengths of the extracellular P-loops and unique sequence motifs. The 3JBR analysis, which also concluded that the repeats arrange in a clockwise orientation around the pore as viewed from the extracellular surface, matched experimental results (electron density in the cryo-EM map) to the following major observations: (1) the loop between S5 and the P1 helix (referred to as L5) in repeats I and III is longer than the L5 loops in repeats II and IV; (2) repeat III contains a unique YYYVY repeat in L5 that distinguishes repeat I from repeat III; and (3) the length between the P2 helix and S6 (L6) in repeat IV is longer than repeat II. We independently confirmed that, indeed, (1) L5 is longer for repeat I (57 residues) and repeat III (47 residues) than for repeats II (19 residues) and IV (18 residues); (2) repeat III contained the YYYVY motif reported (located at Y971): and (3) L6 was longer in repeat IV (35 residues) than in repeat II (21 residues). Correlating observations from two independent cryo-EM reports with our bioinformatics analysis and inspection of the electron density data confirmed a clockwise orientation of the repeats around the pore as viewed from the extracellular surface and as reported for human Cav3.1 (Walsh et al., 2009) and later for rabbit Cav1.1. The PDB: 5GJV model further confirmed this arrangement of the repeats.
The only structural feature not recapitulated in our 3RVY-R model and reliably represented in the PDB: 5GJV map is the reported 30° angle deviation in the spatial orientation of the four-helix bundle of repeats II and III, which likely represents biologically relevant, accurate structural information uniquely provided by the cryo-EM maps and raw data. To account for this unique finding, we used the 3RVY-R model to correct the inaccuracies in residue assignment of S3 found in repeats I and III of PDB: 5GJV by iteratively globally superimposing the Cα carbons of the PDB: 5GJV and 3RVY-R models to minimize the RMSD between them and then using the superimposition to guide manual modifications in the sequence alignment, which clearly introduced gaps near areas of misalignment. A homology model was then built using the coordinates of the PDB: 5GVJ cryo-EM-based model as a template but reassigning the residues to their correct position using the residue assignment of the 3RVY-R model as guide. This PDB: 3RVY-”corrected 5GJV” model restores the motif in the two repeats where they are absent (I and III) by moving the relevant side chains, which are conserved throughout the family, to their expected location, where they are positioned to the same face of the 310 alpha helix in S3, facing toward the hydrophobic core of the VSD four-helix bundle in all segments, and corresponding to the equivalent turn of the helix within the α helix in S3 as in the original PDB: 3RVY crystal structure, which also correlates with the other two repeats of PDB: 5GJV (II and IV). The “3RVY-corrected-5GJV” model accurately fits the cryo-EM map at the side-chain density level, with the improvement that our method accounts for conservation of functional inter-domain contacts, which, in this case, translates into accurately modeling all functional residues of the VSD within the structure.
To compare the accuracy of fitness, we performed rigid body fitting of the superimposition of the PDB: 5GJV cryo-EM-based model and our 3RVY-R and corrected PDB: 5GJV homology models onto the PDB: 5GJV cryo-EM map with the Chimera software as depicted in Figure 2.
Human Cav Channel Modeling
To model by homology the α1 subunit of the human Cav family, we first corrected the cryo-EM-based models and then used our corrected model of rabbit Cav1.1 as a structural template to individually construct the subunits by modeling the repeats in a clockwise orientation around the pore as viewed from the extracellular side.
Mapping the Location of Disease-Causing Mutations and Functional Residues
The 3D location of known disease-causing mutations and the functional residues associated with the salt bridge network of the VSD and the selectivity filter sites were analyzed with multiple sequence alignment to identify equivalent amino acids between all human Cav channels, paired with visual inspection andanalysis of the structural location of theside chains of the functional amino acids of interest. To map the structural location of the mutations in the figures, we segmented each of the four repeats of all 10 channels (40 sequences total, 10 sequences per repeat) and performed multiple sequence alignment by repeat. We included repeat I of Cav1.1 in all alignments to find structurally equivalent positions between all repeats and then used this “template” repeat to display the relationships between all mutations that target a specific structural site independent of the channel or disease to which it relates.
Pore Architecture Conservation Analysis
To analyze architectural features of the selectivity filter, we identified equivalent amino acids between all human Cav channels and the engineered CavAb channel with functional mapping as described above (Tang et al., 2014) After identifying the equivalent functional amino acids of CavAb in our homology model of Cav1.1, we proceeded to perform iterative rounds of energy minimization of the side chains of the residues of the selectivity filter and visually analyzed the resultant conformations.
DATA AND SOFTWARE AVAILABILITY
The UniProt ID for the rabbit Cav1.1 sequence used for our 3RVY-R and corrected 5GJV model was P07923. The UniProt ID for the human Cav sequences modeled were as follows: Cav1.1, Q13698; Cav1.2, Q13936; Cav1.3, Q01668; Cav1.4, O60840; Cav2.1, O00555; Cav2.2, Q00975; Cav2.3, Q15878; Cav3.1, O43497; Cav3.2, O95180; and Cav3.3, Q9P0X4. The cryo-EM map used for modeling was EMDB: EM9513 (associated with PDB: 5GJV). The human models are available for download at https://github.com/CardozoLab/Voltage-Gated-Calcium-Channels-/blob/master/README.md. The coordinates of the corrected 5GJV rabbit Cav1.1 α1 model reported in this paper are available at PDB: 6BYO.
Supplementary Material
Highlights.
Core residue contact surface area is quantitative structure accuracy metric
Refine low-resolution X-ray and cryo-EM structures to atomic resolution with metric
Identify/correct inaccuracies in mammalian voltage-gated ion channel structure
Metric-refined channel models illuminate mechanism and disease-causing mutations
Acknowledgments
This work was supported by NIH grants F31HL124898-02 (to W.M.O.) and DP2 OD004631 (to T.C.), as well as institutional funds. We thank Rodolfo Llinas and David Stokes for their intellectual contributions as well as Evgeny Shmelkov for technical support.
Footnotes
Data and Software Availability
6BYO
Supplemental Information includes three figures and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.04.024.
AUTHOR CONTRIBUTIONS
W.M.O. was responsible for study conception and design, performance of experiments, organization and analysis of data, and manuscript writing. T.C. was responsible for study conception and design, organization and analysis of data, and manuscript writing.
DECLARATION OF INTERESTS
The authors declare no financial interests.
References
- Abagyan RA, Batalov S. Do aligned sequences share the same fold? J Mol Biol. 1997;273:355–368. doi: 10.1006/jmbi.1997.1287. [DOI] [PubMed] [Google Scholar]
- Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- Abagyan RA, Totrov MM. Contact area difference (CAD): a robust measure to evaluate accuracy of protein models. J Mol Biol. 1997;268:678–685. doi: 10.1006/jmbi.1997.0994. [DOI] [PubMed] [Google Scholar]
- Adams PJ, Snutch TP. Calcium channelopathies: voltage-gated calcium channels. Subcell Biochem. 2007;45:215–251. doi: 10.1007/978-1-4020-6191-2_8. [DOI] [PubMed] [Google Scholar]
- Bech-Hansen NT, Naylor MJ, Maybaum TA, Pearce WG, Koop B, Fishman GA, Mets M, Musarella MA, Boycott KM. Loss-of-function mutations in a calcium-channel alpha1-subunit gene in Xp11.23 cause incomplete X-linked congenital stationary night blindness. Nat Genet. 1998;19:264–267. doi: 10.1038/947. [DOI] [PubMed] [Google Scholar]
- Caciotti A, Morrone A, Domenici R, Donati MA, Zammarchi E. Severe prognosis in a large family with hypokalemic periodic paralysis. Muscle Nerve. 2003;27:165–169. doi: 10.1002/mus.10298. [DOI] [PubMed] [Google Scholar]
- Cardozo T, Totrov M, Abagyan R. Homology modeling by the ICM method. Proteins. 1995;23:403–414. doi: 10.1002/prot.340230314. [DOI] [PubMed] [Google Scholar]
- Catterall WA, Wisedchaisri G, Zheng N. The chemical basis for electrical signaling. Nat Chem Biol. 2017;13:455–463. doi: 10.1038/nchembio.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Heckenlively JR, Bayley PR, Brecha NC, Davisson MT, Hawes NL, Hirano AA, Hurd RE, Ikeda A, Johnson BA, et al. The nob2 mouse, a null mutation in Cacna1f: anatomical and functional abnormalities in the outer retina and their consequences on ganglion cell visual responses. Vis Neurosci. 2006;23:11–24. doi: 10.1017/S095252380623102X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lu J, Pan H, Zhang Y, Wu H, Xu K, Liu X, Jiang Y, Bao X, Yao Z, et al. Association between genetic variation of CACNA1H and childhood absence epilepsy. Ann Neurol. 2003;54:239–243. doi: 10.1002/ana.10607. [DOI] [PubMed] [Google Scholar]
- Denier C, Ducros A, Vahedi K, Joutel A, Thierry P, Ritz A, Castelnovo G, Deonna T, Gérard P, Devoize JL, et al. High prevalence of CACNA1A truncations and broader clinical spectrum in episodic ataxia type 2. Neurology. 1999;52:1816–1821. doi: 10.1212/wnl.52.9.1816. [DOI] [PubMed] [Google Scholar]
- Dunn SD, Wahl LM, Gloor GB. Mutual information without the influence of phylogeny or entropy dramatically improves residue contact prediction. Bioinformatics. 2008;24:333–340. doi: 10.1093/bioinformatics/btm604. [DOI] [PubMed] [Google Scholar]
- Fouad G, Dalakas M, Servidei S, Mendell JR, Van den Bergh P, Angelini C, Alderson K, Griggs RC, Tawil R, Gregg R, et al. Genotype-phenotype correlations of DHP receptor alpha 1-subunit gene mutations causing hypokalemic periodic paralysis. Neuromuscul Disord. 1997;7:33–38. doi: 10.1016/s0960-8966(96)00401-4. [DOI] [PubMed] [Google Scholar]
- Guda P, Bourne PE, Guda C. Conserved motifs in voltage-sensing and pore-forming modules of voltage-gated ion channel proteins. Biochem Biophys Res Commun. 2007;352:292–298. doi: 10.1016/j.bbrc.2006.10.190. [DOI] [PubMed] [Google Scholar]
- Heron SE, Phillips HA, Mulley JC, Mazarib A, Neufeld MY, Berkovic SF, Scheffer IE. Genetic variation of CACNA1H in idiopathic generalized epilepsy. Ann Neurol. 2004;55:595–596. doi: 10.1002/ana.20028. [DOI] [PubMed] [Google Scholar]
- Jen J, Yue Q, Nelson SF, Yu H, Litt M, Nutt J, Baloh RW. A novel nonsense mutation in CACNA1A causes episodic ataxia and hemiplegia. Neurology. 1999;53:34–37. doi: 10.1212/wnl.53.1.34. [DOI] [PubMed] [Google Scholar]
- Jeng CJ, Chen YT, Chen YW, Tang CY. Dominant-negative effects of human P/Q-type Ca2+ channel mutations associated with episodic ataxia type 2. Am J Physiol Cell Physiol. 2006;290:C1209–C1220. doi: 10.1152/ajpcell.00247.2005. [DOI] [PubMed] [Google Scholar]
- Jurkat-Rott K, Lehmann-Horn F, Elbaz A, Heine R, Gregg RG, Hogan K, Powers PA, Lapie P, Vale-Santos JE, Weissenbach J, et al. A calcium channel mutation causing hypokalemic periodic paralysis. Hum Mol Genet. 1994;3:1415–1419. doi: 10.1093/hmg/3.8.1415. [DOI] [PubMed] [Google Scholar]
- Jurkat-Rott K, Lerche H, Lehmann-Horn F. Skeletal muscle channelopathies. J Neurol. 2002;249:1493–1502. doi: 10.1007/s00415-002-0871-5. [DOI] [PubMed] [Google Scholar]
- Kamisetty H, Ovchinnikov S, Baker D. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc Natl Acad Sci USA. 2013;110:15674–15679. doi: 10.1073/pnas.1314045110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravani H, Bladen C, Parker DB, Snutch TP, McRory JE, Zamponi GW. Effects of Cav3.2 channel mutations linked to idiopathic generalized epilepsy. Ann Neurol. 2005;57:745–749. doi: 10.1002/ana.20458. [DOI] [PubMed] [Google Scholar]
- Mantuano E, Veneziano L, Spadaro M, Giunti P, Guida S, Leggio MG, Verriello L, Wood N, Jodice C, Frontali M. Clusters of non-truncating mutations of P/Q type Ca2+ channel subunit Ca(v)2.1 causing episodic ataxia 2. J Med Genet. 2004;41:e82. doi: 10.1136/jmg.2003.015396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRory JE, Hamid J, Doering CJ, Garcia E, Parker R, Hamming K, Chen L, Hildebrand M, Beedle AM, Feldcamp L, et al. The CACNA1F gene encodes an L-type calcium channel with unique biophysical properties and tissue distribution. J Neurosci. 2004;24:1707–1718. doi: 10.1523/JNEUROSCI.4846-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ophoff RA, Terwindt GM, Vergouwe MN, van Eijk R, Oefner PJ, Hoffman SM, Lamerdin JE, Mohrenweiser HW, Bulman DE, Ferrari M, et al. Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell. 1996;87:543–552. doi: 10.1016/s0092-8674(00)81373-2. [DOI] [PubMed] [Google Scholar]
- Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486:135–139. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptácek LJ, Tawil R, Griggs RC, Engel AG, Layzer RB, Kwieciński H, McManis PG, Santiago L, Moore M, Fouad G, et al. Dihydropyridine receptor mutations cause hypokalemic periodic paralysis. Cell. 1994;77:863–868. doi: 10.1016/0092-8674(94)90135-x. [DOI] [PubMed] [Google Scholar]
- Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- Shen H, Zhou Q, Pan X, Li Z, Wu J, Yan N. Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science. 2017;355:eaal4326. doi: 10.1126/science.aal4326. [DOI] [PubMed] [Google Scholar]
- Spacey SD, Hildebrand ME, Materek LA, Bird TD, Snutch TP. Functional implications of a novel EA2 mutation in the P/Q-type calcium channel. Ann Neurol. 2004;56:213–220. doi: 10.1002/ana.20169. [DOI] [PubMed] [Google Scholar]
- Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, Napolitano C, Schwartz PJ, Joseph RM, Condouris K, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Splawski I, Timothy KW, Decher N, Kumar P, Sachse FB, Beggs AH, Sanguinetti MC, Keating MT. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc Natl Acad Sci USA. 2005;102:8089–8096. doi: 10.1073/pnas.0502506102. discussion 8086–8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splawski I, Yoo DS, Stotz SC, Cherry A, Clapham DE, Keating MT. CACNA1H mutations in autism spectrum disorders. J Biol Chem. 2006;281:22085–22091. doi: 10.1074/jbc.M603316200. [DOI] [PubMed] [Google Scholar]
- Striessnig J, Hoda JC, Koschak A, Zaghetto F, Mullner C, Sinnegger-Brauns MJ, Wild C, Watschinger K, Trockenbacher A, Pelster G. L-type Ca2+ channels in Ca2+ channelopathies. Biochem Biophys Res Commun. 2004;322:1341–1346. doi: 10.1016/j.bbrc.2004.08.039. [DOI] [PubMed] [Google Scholar]
- Strom TM, Nyakatura G, Apfelstedt-Sylla E, Hellebrand H, Lorenz B, Weber BH, Wutz K, Gutwillinger N, Ruther K, Drescher B, et al. An L-type calcium-channel gene mutated in incomplete X-linked congenital stationary night blindness. Nat Genet. 1998;19:260–263. doi: 10.1038/940. [DOI] [PubMed] [Google Scholar]
- Tang L, Gamal El-Din TM, Payandeh J, Martinez GQ, Heard TM, Scheuer T, Zheng N, Catterall WA. Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature. 2014;505:56–61. doi: 10.1038/nature12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tress ML, Valencia A. Predicted residue-residue contacts can help the scoring of 3D models. Proteins. 2010;78:1980–1991. doi: 10.1002/prot.22714. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Tani K, Irie K, Hiroaki Y, Shimomura T, McMillan DG, Cook GM, Schertler GF, Fujiyoshi Y, Li XD. Two alternative conformations of a voltage-gated sodium channel. J Mol Biol. 2013;425:4074–4088. doi: 10.1016/j.jmb.2013.06.036. [DOI] [PubMed] [Google Scholar]
- Van Petegem F, Minor DL., Jr The structural biology of voltage-gated calcium channel function and regulation. Biochem Soc Trans. 2006;34:887–893. doi: 10.1042/BST0340887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitko I, Chen Y, Arias JM, Shen Y, Wu XR, Perez-Reyes E. Functional characterization and neuronal modeling of the effects of childhood absence epilepsy variants of CACNA1H, a T-type calcium channel. J Neurosci. 2005;25:4844–4855. doi: 10.1523/JNEUROSCI.0847-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace SJ. Use of ethosuximide and valproate in the treatment of epilepsy. Neurol Clin. 1986;4:601–616. [PubMed] [Google Scholar]
- Walsh CP, Davies A, Butcher AJ, Dolphin AC, Kitmitto A. Three-dimensional structure of CaV3.1: comparison with the cardiac L-type voltage-gated calcium channel monomer architecture. J Biol Chem. 2009;284:22310–22321. doi: 10.1074/jbc.M109.017152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wappl E, Koschak A, Poteser M, Sinnegger MJ, Walter D, Eberhart A, Groschner K, Glossmann H, Kraus RL, Grabner M, Striessnig J. Functional consequences of P/Q-type Ca2+ channel Cav2.1 missense mutations associated with episodic ataxia type 2 and progressive ataxia. J Biol Chem. 2002;277:6960–6966. doi: 10.1074/jbc.M110948200. [DOI] [PubMed] [Google Scholar]
- Wu J, Yan Z, Li Z, Yan C, Lu S, Dong M, Yan N. Structure of the voltage-gated calcium channel Cav1.1 complex. Science. 2015;350:aad2395. doi: 10.1126/science.aad2395. [DOI] [PubMed] [Google Scholar]
- Wu J, Yan Z, Li Z, Qian X, Lu S, Dong M, Zhou Q, Yan N. Structure of the voltage-gated calcium channel Ca(v)1.1 at 3.6 Å resolution. Nature. 2016;537:191–196. doi: 10.1038/nature19321. [DOI] [PubMed] [Google Scholar]
- Yu FH, Catterall WA. The VGL-chanome: a protein superfamily specialized for electrical signaling and ionic homeostasis. Sci STKE. 2004;2004:re15. doi: 10.1126/stke.2532004re15. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ren W, DeCaen P, Yan C, Tao X, Tang L, Wang J, Hasegawa K, Kumasaka T, He J, et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486:130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.