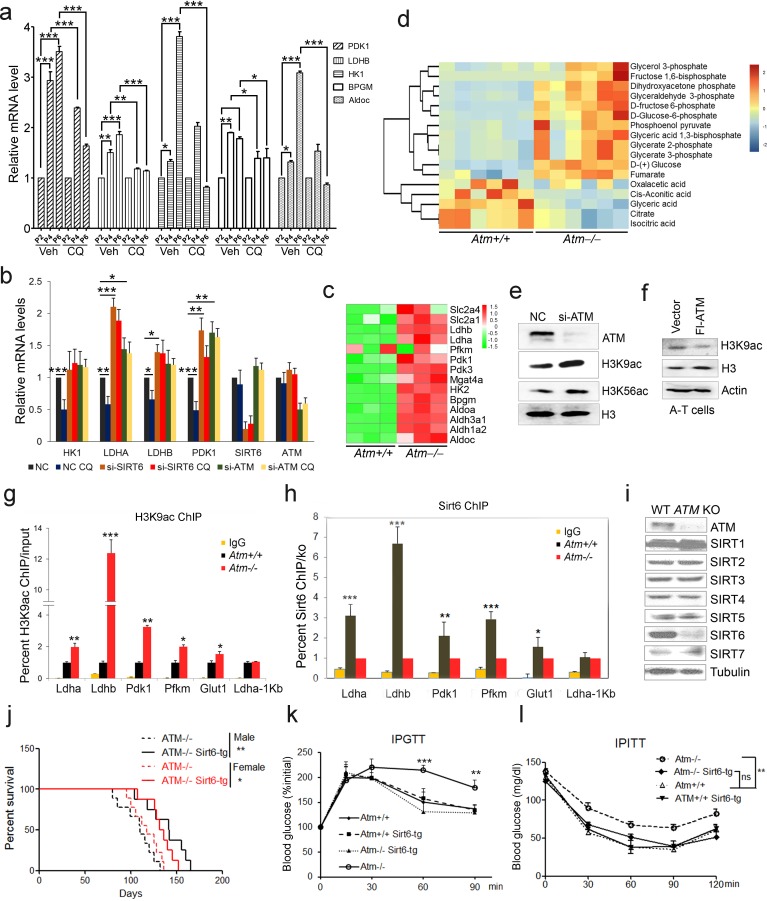
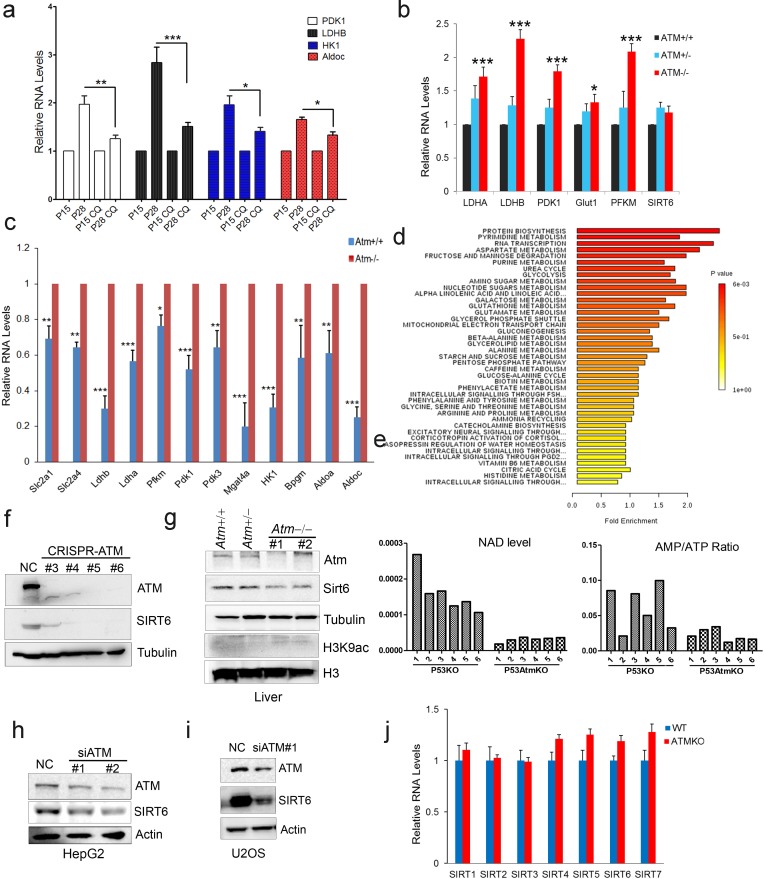
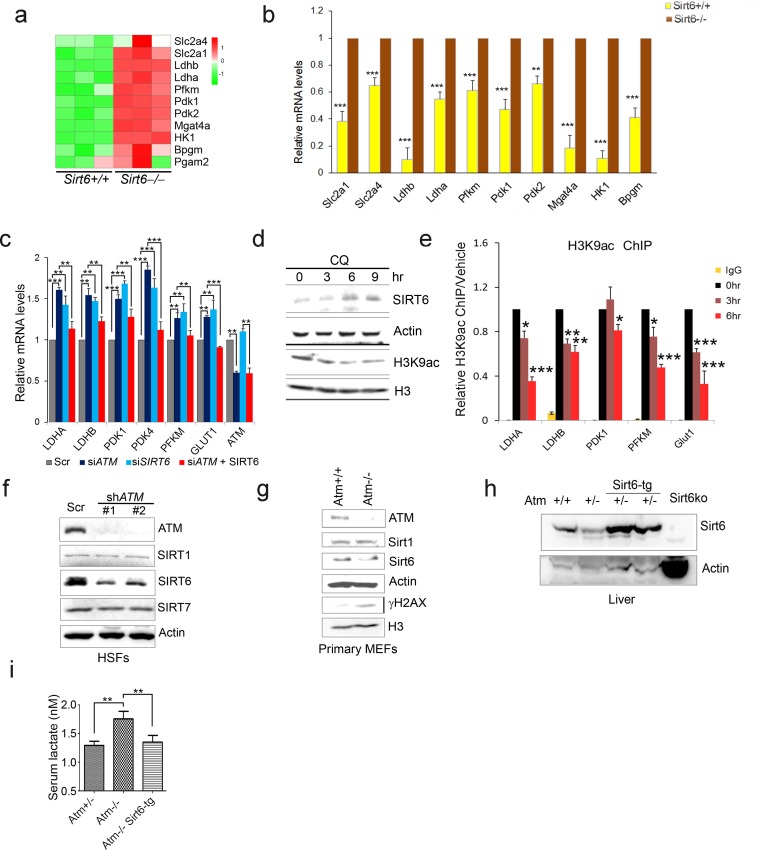
Figure 2. ATM-SIRT6 axis regulates age-related metabolic reprogramming.
(a) Quantitative RT-PCR analysis of mRNA levels of indicated glycolytic genes in different passages of MEFs with or without treatment of CQ. Data represent means ± SEM. *p<0.05, **p<0.01, ***p<0.001. (b) Quantitative RT-PCR analysis of mRNA levels of indicated glycolytic genes in Scramble (NC), si-SIRT6 or si-ATM HepG2 cells incubated with or without CQ (10 μM, 6 hr). Data represent means ± SEM. *p<0.05, **p<0.01, ***p<0.001. (c) Heatmap representation of RNA-Seq data (GSE109280) showing relative changes of glycolytic genes in Atm-/- MEF cells. The transcript levels are qualified in reads per kilobase of exon per million mapped sequence reads (RPKM), which is a normalized measure of exonic read density. Red and green indicate up- and downregulation, respectively. (d) Heatmap showing relative levels of metabolites in Atm+/+ and Atm-/- MEF cells of p53 null background, analyzed by LC-MS. Red and blue indicate up- and downregulation, respectively. (e) Immunoblots showing protein levels of H3K9ac and H3K56ac in ATM-deficient HepG2 cells. (f) Immunoblots showing levels of H3K9ac in A-T cells reconstituted with Flag-ATM. (g) ChIP analysis showing enrichment of H3K9ac at the promoter regions of indicated genes in Atm+/+ and Atm-/- MEFs. Data represent means ± SEM of three independent experiments. *p<0.05, **p<0.01, ***p<0.001. (h) ChIP analysis showing enrichment of Sirt6 at the promoter regions of indicated genes in Atm+/+ and Atm-/- MEFs. Data represent means ± SEM of six independent experiments. *p<0.05, **p<0.01, ***p<0.001. (i) Immunoblots showing protein levels of sirtuins in wild-type (WT) and ATM knockout (KO) HEK293 cells. (j) Kaplan-Meier survival of Atm-/- and Atm-/-;Sirt6-Tg male (n = 11 in each group) and female (n = 9 in each group) mice. **p<0.01. (k) Results of glucose tolerance tests in Atm+/+, Atm-/-, and Atm-/-;Sirt6-Tg mice. Data represent means ± SEM, n = 6. **p<0.01, ***p<0.001. (l) Results of insulin tolerance tests in Atm+/+, Atm-/-, and Atm-/-;Sirt6-Tg mice. Data represent means ± SEM, n = 6. **p<0.01. ‘ns’ indicates no significant difference.