Figure 2. KLF4-mCpG binding activity activates genes involved in GBM cytoskeletal organization and migration.
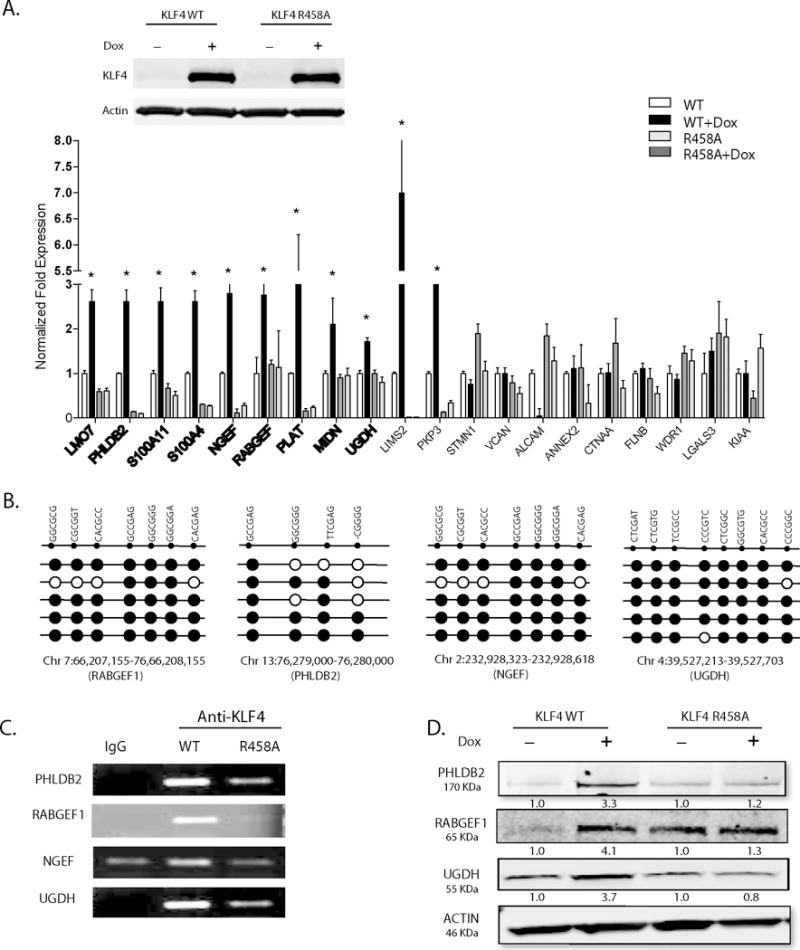
(A) Upper panel: Western blot analysis showed KLF4 WT or KLF4 R458A expression in tet-on stable U87 GBM cell lines upon doxycyline treatment (1 ug/ml, 48 hrs). Lower panel: Twenty putative KLF4-mCpG gene targets involved in cell migration pathway were picked from our previous RNA-seq studies. Real time-PCR (RT-PCR) revealed 11 of the 20 genes were significantly upregulated by KLF4 WT only, with no change in KLF4 R458A expressing cells (+Dox, 48 hr), confirming a mCpG-dependent gene activation mechanism (asterisks). Bold showed targets with methylated cis-regulatory elements in the gene. (B) Sanger bisulfite sequencing indicated DNA methylation in tested cis-regulatory regions of putative KLF4-mCpG targets. Examples of four genes showed highly methylated KLF4 binding regions of these genes. Each row represents one sequenced clone; each column represents one CpG site; filled circles stand for methylation. (C) Confirmation that KLF4 WT but not KLF4 R458R preferentially bound to the methylated cis-regulatory regions of selected genes. A KLF4 antibody was used to precipitate cross-linked genomic DNA from U87 cells expressing KLF4 WT or KLF4 R458A. Rabbit IgG was used to control for non-specific binding. De-crosslinked DNA samples were served as the input for ChIP-PCR. KLF4 binding to the selected regions was enriched in KLF4 WT expressing cells. (D) Western blot analysis indicating increased protein expression of the selected targets by KLF4 WT, but not by KLF4 R458A.
