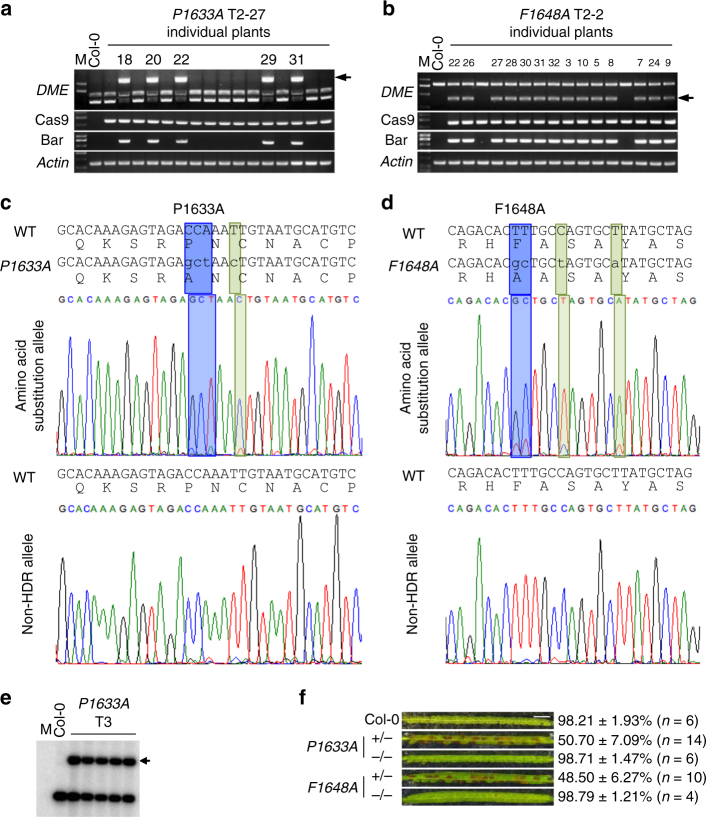
Fig. 4.
Single amino acid substitution at the endogenous DME locus by gene targeting. a, b Genotyping of T2 individual plants for P1633A and F1648A, respectively. Arrows indicate the specific sequence substitution as detected by PCR and restriction enzyme digestion. PCR primers are as depicted in Supplementary Figure 6a and Supplementary Table 4. Detailed information is described in Supplementary Figure 6. c, d Sequence confirmation of the P1633A and F1648A substitutions, respectively, in individual T2 plants. Blue highlights indicate amino acid substitution, green highlights indicate silent mutations. The sequence chromatograms are taken from FinchTV. e Southern blotting. The P1633A substitutions were confirmed by Southern blotting in heterozygous T3 plants. The arrow indicates the band caused by specific base substitution. f Analysis of seed abortion. Seeds from Col-0, heterozygous GT (+/−), and non-GT (−/−) P1633A and F1648A T3 plants were analyzed. Scale bar, 1 mm