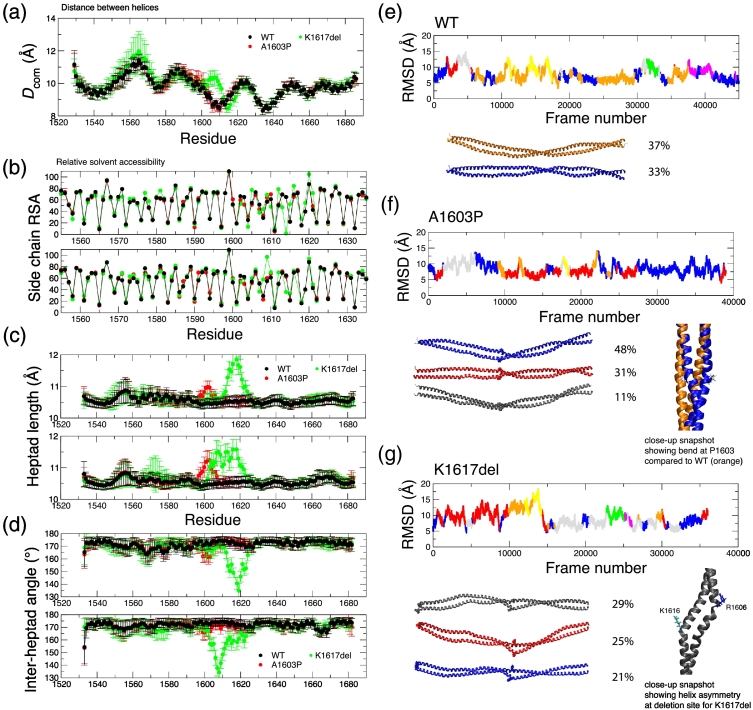
Fig. 8.
Results from MD simulation of the composite model coiled-coil and mutant models. (a) The distance between the helices (Dcom) as a function of residue position for WT (black), A1603P (red) and K1617del (green). Error bars represent the SD during the simulation. (b) Relative solvent accessibility (RSA) for side-chain atoms in each residue. Low values indicate solvent exclusion and thereby highlight the core residues; upper and lower plots show the RSA values for chains A and B, respectively; color scheme as per part a. Error bars are omitted for clarity. (c) Heptad length, the distance between Cα atoms in residues at positions i and i + 7, along chain A (upper) and chain B (lower). (d) Inter-heptad angle, the angle between the lines linking Cα atoms in residues at positions (i – 7), (i) and (i, i + 7), for chain A (upper) and chain B (lower). RMSD for all Cα atoms compared to the initial structure for WT (e), A1603P (f), and K1617del (g). The plots are divided by color into the different structure clusters. Example structures for the most heavily populated clusters (> 10%) are shown beneath. The position of the residue being investigated (or K1616 in the case of K1617del) is shown in spacefill. The cluster populations as a percentage of all trajectory snapshot structures are indicated. Inset for A1603P: close-up snapshot images of representative structures from the most heavily populated clusters in A1603P (blue, showing the bend at P1603) compared to WT (orange). Inset for K1617del: close-up snapshot image of the representative structure from the most heavily populated cluster showing helix asymmetry at the deletion site with bends appearing near R1606 in helix B (blue, right) and near K1616 in helix A (cyan, left).