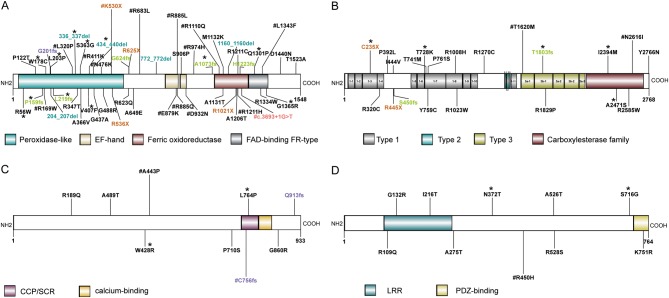
Figure 3.
Mutation sites in the secondary structure of DUOX2, TG, TSHR and TPO proteins. (A) Fifty-one mutation sites distributed in the secondary structure of DUOX2 protein, which contains 4 domains, peroxidase-like domain, EF-hand domain, ferric oxidoreductase domain and FAD-binding FR-type domain. A total of 41 mutations were located in these domains region. (B) Twenty-one mutation sites located in the secondary structure of TG protein, which include 3 domains, type 1 domain, type 2 domain, type 3 domain and carboxylesterase family domain. Fifteen mutation sites were in domain region. (C) Nine mutation sites were in TPO protein with CCP/SCR domain and calcium domain, only two mutations (L764P, C756fs) were in domain region. (D) Ten mutation sites were located in TSHR protein, of which 3 were in LRR domain and one was in PDZ-binding domain. FAD, flavin adenine dinucleotide; CCP, the complement control protein; SCR, short consensus repeat; LRR, leucine-rich repeats. Missense mutations, stopgain mutations, splicing mutations, frameshift deletion mutations, frameshift insertion mutations and nonframeshift deletion mutations are indicated with black, brown, red, green, purple and blue font, respectively. *Denotes the novel mutations and #denotes the recurrent mutations.

 This work is licensed under a
This work is licensed under a