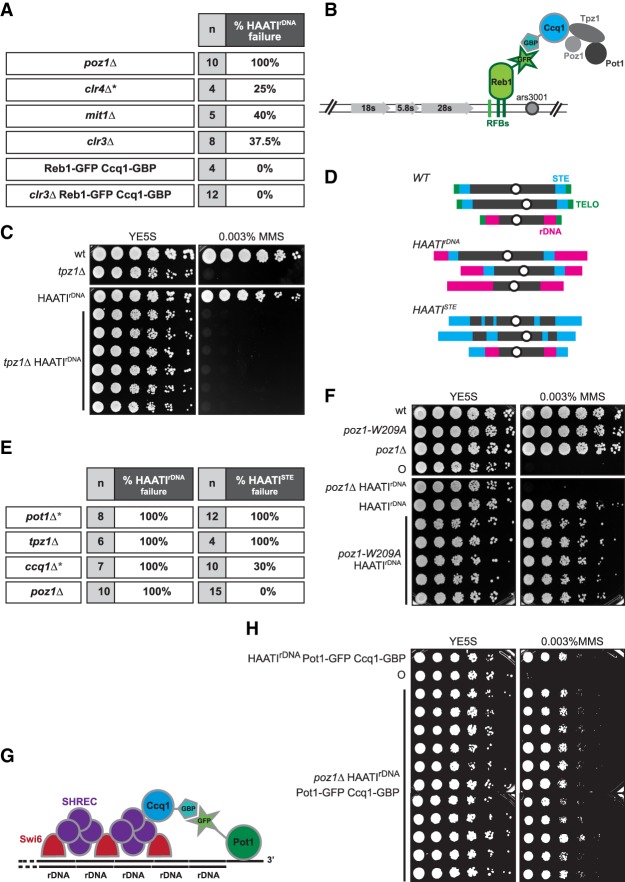
Figure 1.
Pot1 engagement at HAATI chromosome ends. (A) Table showing the requirement of shelterin and SHREC proteins for maintenance of HAATIrDNA chromosomes. The gene deletions indicated in the table (poz1Δ, mit1Δ, and clr3Δ) were constructed by one-step gene replacement in already formed haploid HAATIrDNA survivors. Previously reported data for clr4+ deletion (Jain et al. 2010) are marked by an asterisk and shown for comparison. “n” indicates the total number of HAATIrDNA transformants analyzed for each gene deletion; percentages indicate the frequency at which HAATIrDNA capability is lost, leading to chromosome circularization. This was scored first by exposure to MMS, as circulars display extreme hypersensitivity to this agent (Jain et al. 2010); for a subset of isolates, the chromosome circularity was further confirmed by PFGE (explained in detail in Fig. 3). (B) Schematic illustrating the strategy for tethering Ccq1 to rDNA arrays in the absence of SHREC by forcing interaction between Reb1 and Ccq1 via the GFP-binding protein (GBP)–GFP interaction. (C) Fivefold serial dilutions of the indicated (107 cells per milliliter) cultures on medium lacking or containing MMS. (D) Schematic depicting wild-type and HAATI chromosome organization. (Top) In wild-type cells, the telomeres on chromosomes I and II are flanked by the STE regions, comprising ∼20 kb of imperfect ∼86-bp repeats. The telomeres of chromosome III are flanked by the rDNA repeats, which comprise ∼1 Mb of the 3.6-Mb chromosome. (Middle) HAATIrDNA contains rDNA at the ends of all chromosomes. (Bottom) In HAATISTE cells, STE sequences localize to the ends of all chromosomes as well as multiple sites in the chromosomal interiors. (E) Table summarizing the requirement of telomere-associated proteins for maintenance of HAATIrDNA versus HAATISTE. The genes indicated in the table were deleted by one-step gene replacement in already formed haploid HAATIrDNA or HAATISTE survivors. Percentages indicate the frequency of chromosome circularization in each HAATI subtype. Chromosome circularization was measured by assessing MMS sensitivity. The asterisks mark published results (Jain et al. 2010) shown for comparison. (F) Dilution assay performed as in C. (G) Schematic illustrating the strategy for tethering Pot1 to Ccq1 (via GBP–GFP interaction) in the absence of Poz1. (H) Dilution assay performed as in C. (○) Circular strain.