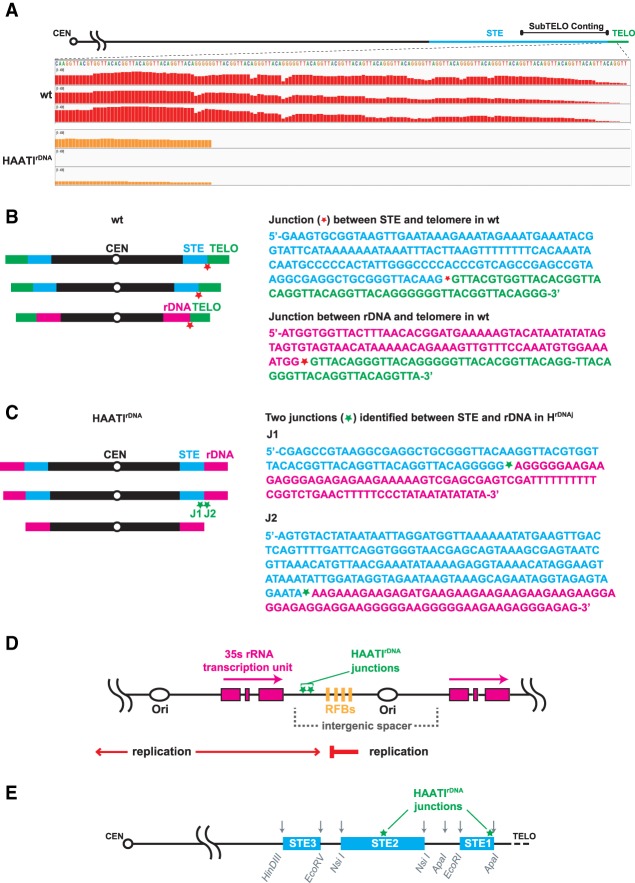
Figure 6.
Whole-genome sequencing of HAATIrDNA isolates identifies junctions derived from rDNA translocation. (A) Screenshot of Integrated Genomic Viewer browser depicting paired-end sequencing reads mapped to the subtelomeric (subTELO) contig, which harbors 7848 bp of STE DNA and 202 bp of telomere repeats (see the Materials and Methods). Three replicates of wild-type cells show robust mapping of reads to the telomeric repeats; the near-complete absence of corresponding reads in HAATIrDNA isolates confirms the absence of canonical telomeres. The few sequencing reads culminating in telomeric repeats in HAATIrDNA correspond to the four telomere repeat units found within the STE region centromere-proximal to the translocation site in a subset of HAATI isolates. The scale for bar height in both wild-type and HAATIrDNA samples is 0–400 and indicates the relative level of coverage of each base in a DNA string. (B, left) Wild-type genome organization. (Right) Sequences just centromere-proximal to the telomeric repeats on each chromosome, as identified by whole-genome sequencing from three wild-type isolates, are shown. Red asterisks denote the junctions between wild-type telomeric and STE (top sequence) or rDNA (bottom sequence) regions. (C, left) HAATIrDNA genome organization. (Right) Unique junctions (J1 and J2) between the centromere-proximal STE regions and rDNA in HAATIrDNA genomes. Green asterisks denote the junctions, which are unique to HAATIrDNA strains. (D) Schematic of the rDNA region. Each 10.9-kb rDNA repeat is composed of the 35S rDNA transcriptional unit (pink boxes) and an intergenic spacer that includes one origin of replication (ars3001) and four closely spaced polar replication fork barriers (RFBs). Green asterisks within the intergenic spacer indicate the sites of junctions between the STE regions (always on the centromere-proximal side) and the rDNA repeats; these junctions are unique to HAATIrDNA cells. (E) Schematic of the STE regions on chromosome I and chromosome II, which comprise ∼20 kb of imperfect tandem repeats (light-blue boxes) whose sequences modulate from the distal to the proximal ends of the region. Restriction sites (gray arrows) in the STEs are shown. Green asterisks within the STE region indicate the sites of junctions between STEs and rDNA (as indicated by an asterisk in C) in HAATIrDNA.