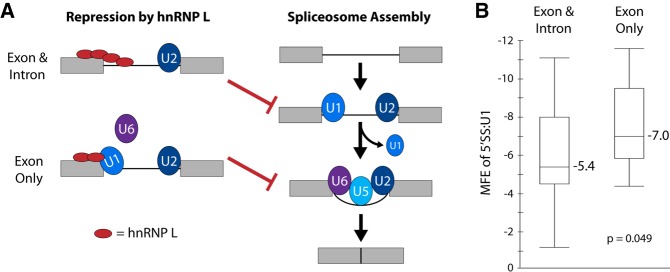
FIGURE 4.
Model for different mechanisms of hnRNP L repression of weak and strong 5′ splice sites. (A) Schematic of normal splicesome assembly (right) and mechanisms by which hnRNP L represses normal assembly. Exons are designated as gray boxes, introns are black lines. The U snRNP components of the spliceosome are indicated by blue and purple ovals. HnRNP L is indicated by a red oval. HnRNP L can either block accessibility of the 5′SS to U1 (exon and intron), or inhibit exchange of U1 for U6 (exon only). In either case, assembly of the final spliceosomal complex is prevented. (B) Minimal free energy (MFE) of the association of the 5′SS with the U1 snRNA for hnRNP L-repressed exons that have CA dinucleotides in the exon and/or intron flanking the 5′SS. See Supplemental Table 1 for details. As described in the text, binding of hnRNP L to the exon only is predicted to repress U1–U6 exchange and is typically marked by a strong 5′SS:U1 interaction. P-value is a two-tailed t-test.