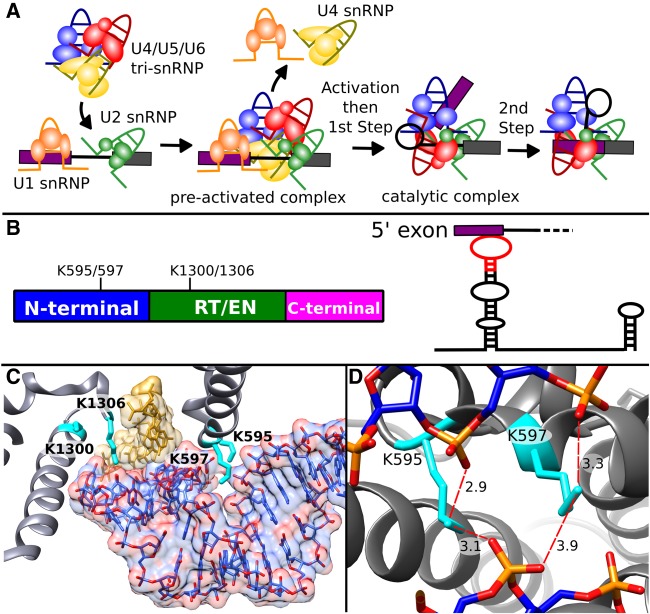
FIGURE 2.
Prp8 lysine residues surrounding U5 snRNA change accessibility between preactivation and catalysis. (A) Schematic of spliceosome assembly highlighting involvement of U snRNPs in the formation of the catalytic core during activation, and the two steps of splicing chemistry. (B, left) Schematic of Prp8 domain structure and relative positions of four differentially modified lysine residues that spatially located near U5 snRNA in the spliceosome. RT/EN indicates the large reverse transcriptase/endonuclease domain. (Right) Secondary structure of U5 snRNA highlighting SLI interaction with the 5′ exon of pre-mRNA. Red indicates the region of U5 snRNA shown in C. (C) Position of Prp8 (gray) side chains K595, K597, K1300, and K1306 (cyan) interactions with U5 SLI (blue) contacting the 5′ exon (gold) U5 SLI heteroatoms are colored red. (D) View of Prp8 K595 and K597 in close proximity to the phosphate backbone of U5 SLI. Distances (Å) from lysine terminal nitrogens and nonbridging oxygens of phosphates of U5 snRNA C45, U46, and A30 are indicated by the dotted red lines. Panels C and D are derived from the cryo-EM model of the human catalytic (C*) spliceosome (Zhang et al. 2017).