FIGURE 3.
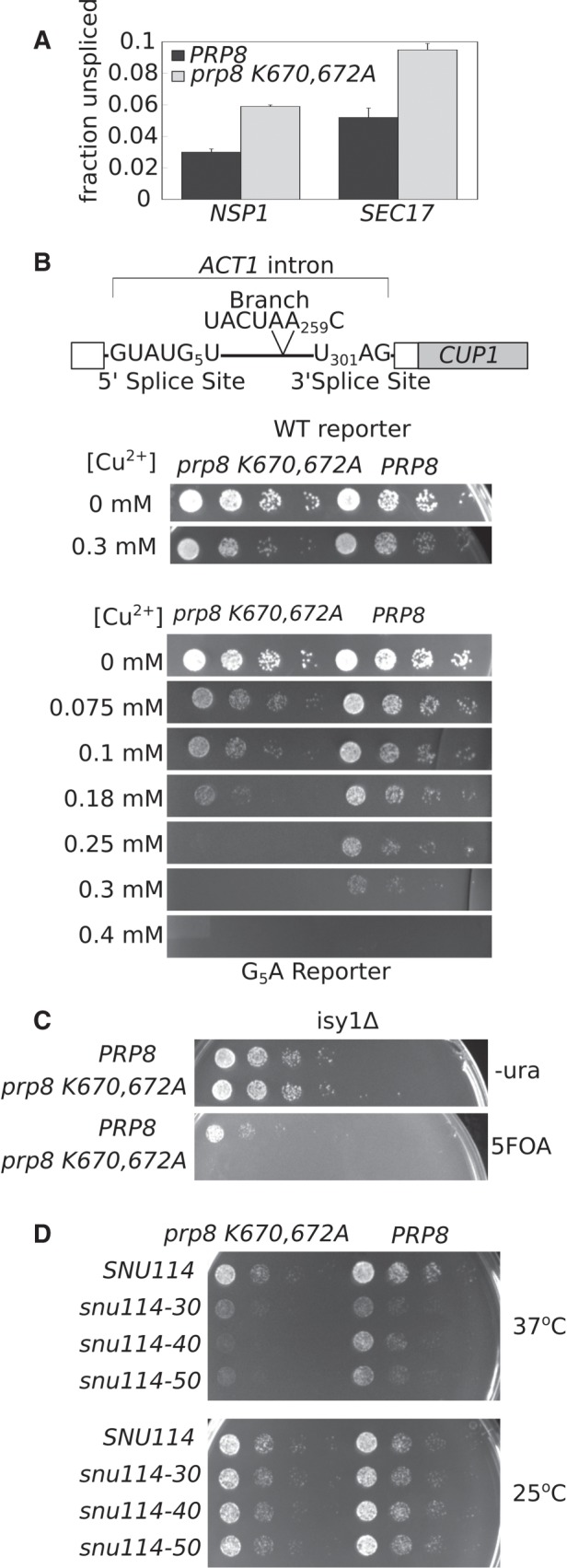
prp8-K670/672A genetically interacts with 5′ splice site and splicing factors involved in activation and first step chemistry. (A) qRT-PCR analysis of relative NSP1 and SEC17 pre-mRNA transcript abundance in prp8-K670/672A (gray bars) and PRP8 (black bars). Primers that span the intron-exon junction were used to quantify pre-mRNA abundance, while primers that amplify a portion of the 3′ exon present in both the pre- and mature mRNA were used to quantify total RNA levels. Fraction unspliced is calculated as Fpre-mRNA/Ftotal. The average of three biological replicates is shown. Error bars represent standard error of the mean. (B) The ACT1–CUP1 splicing reporter. The ACT1 intron is fused to the CUP1 coding sequence such that splicing efficiency can be directly assessed by relative yeast growth on copper-containing media. The 5′ and 3′ and branch site consensus sequences are shown. Numbers indicate position of specific nucleotide where the first nucleotide of the intron is defined as one. Prp8-K670/672A and PRP8 yeast strains carrying either a WT or G5A ACT1–CUP1 reporter were serially diluted and spotted onto plates containing varying concentrations of Cu2+ and allowed to grow at 30°C for 2 d. (C) Double mutant isy1Δ prp8Δ strains carrying either prp8-K670/672A or PRP8 on a HIS plasmid and PRP8 on a URA plasmid were serially diluted and spotted onto 5FOA or –ura plates and grown at 30°C. 5FOA prohibits growth of URA+ strains. prp8-K670/672A is lethal in an isy1Δ background. (D) Double mutant snu114Δ prp8Δ complemented with prp8-K670/672A or PRP8 in combination with SNU114, or snu114-30, snu114-40, or snu114-50 C-terminal truncation alleles were serially diluted and spotted onto rich media and allowed to grow at 37°C and 25°C.
