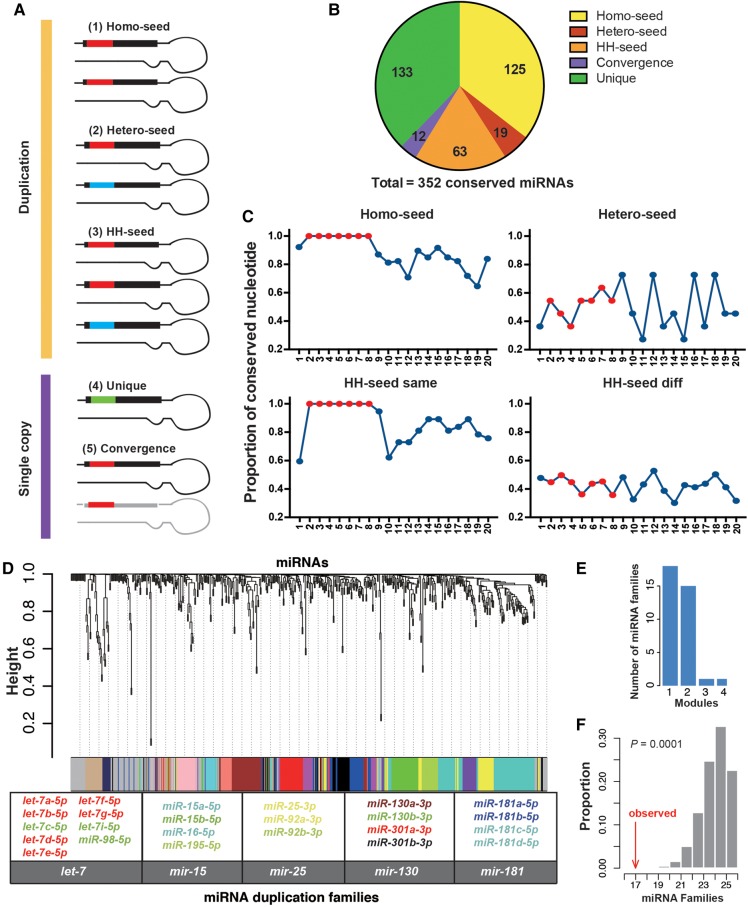
FIGURE 1.
The divergence in sequences and expression patterns between duplicated miRNAs. (A) The classification of human miRNAs that are evolutionarily conserved. Based on the seed sequences of the paralogous miRNAs, the DmiRs are divided into three categories: (1) the homo-seed families; (2) the hetero-seed families; and (3) the homo–hetero-seed (HH-seed) families. For the single-copy mRNAs, they can have seeds identical to other miRNAs that do not have sequence similarity in the precursors due to convergent evolution (Convergence), or they have unique seeds (SCUmiRs). (B) The numbers of miRNA precursors in each of the five categories as described in A. (C) The proportion of nucleotides that are identical between two paralogous miRNAs (y-axis) along the position (x-axis) of the mature miRNAs. On each position, the proportion of the pairwise comparisons that have the same nucleotides out of the total number of pairwise comparisons (y-axis) is given. “HH-seed same,” the paralogous miRNAs that share the same seeds in the HH-seed families; “HH-seed diff,” the paralogous miRNAs that have different seeds in the HH-seed families. Positions 2–8 are shown in red. (D) Hierarchical clustering of 574 miRNAs from 181 nonredundant human tissues/cell lines using WGCNA. The color row below the dendrogram shows the module assignment for each miRNA. The histogram shows the number of miRNA families (y-axis) that had paralogous miRNAs assigned to a single (1, x-axis) module or multiple (2, 3, or 4, x-axis) modules. Five representative miRNA families that had paralogs assigned to at least two different modules are given, with each miRNA member labeled in the same color as the module containing that miRNA. (E) The number of miRNA families (y-axis) that had broadly conserved paralogs assigned to different numbers of modules (x-axis). (F) The observed number of miRNA families that had paralogous copies assigned to at least two different expression modules (the red arrow) and the distribution of the simulated numbers (x-axis) obtained by randomly permuting the miRNA: module assignments for 10,000 replicates (the mean is 25, and 95% CI is [22, 26]).