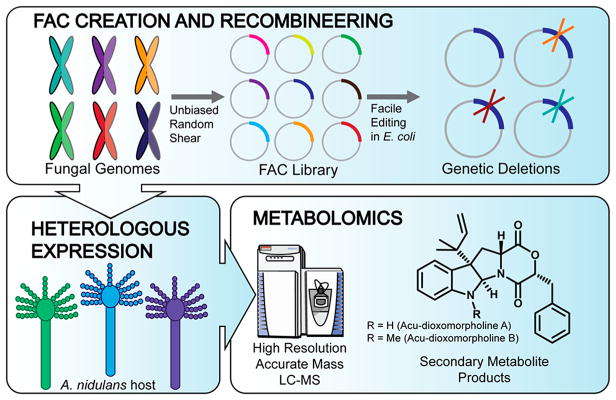
Figure 1.
Platform for discovery of fungal secondary metabolites and their biosynthetic pathways using fungal artificial chromosomes and mass spectrometry-based metabolomic scoring (FAC-MS). Fungal genomes are randomly sheared, and ~100 kb fragments with BGCs are inserted into FACs (top), which are A. nidulans/E. coli shuttle vectors. This enables facile deletion of biosynthetic genes (top, right). FACs are transformed into A. nidulans, a model fungus which serves as a heterologous host (bottom, at left). MS-based metabolomics is utilized to detect secondary metabolites associated with each FAC and to determine the effect of BGC mutants (bottom, at right).