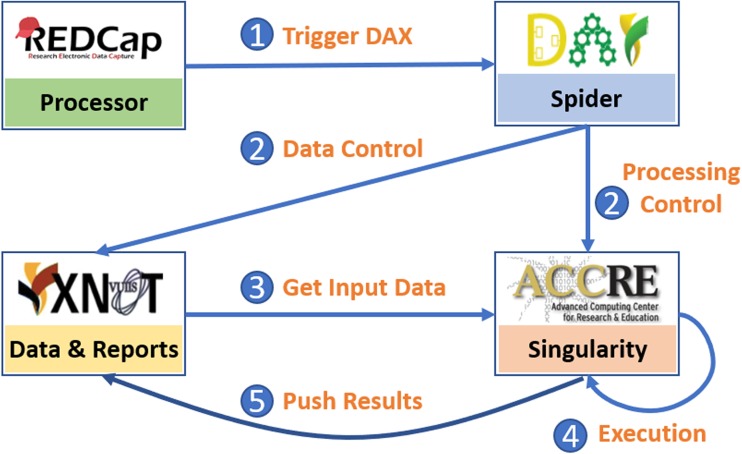
Fig. 4.
The working flow of image processing tasks using VUIIS CCI medical image data storage and processing infrastructure. (1) RedCap triggers DAX to start image processing based on a pre-defined “.yaml” configuration file, specifying which spider to execute (code is available on https://github.com/MASILab/Containerized_DAX). (2) DAX defines data retrieval, computing environment, and software dependencies from a python based spider (code is provided in APPENDIX). (3) Computational infrastructure (e.g., ACCRE) downloads the input data from XNAT. (4) Computational infrastructure executes the related processing algorithms (e.g., “Deep_MultiOrganSeg,” whose Docker image is available on https://hub.docker.com/r/masidocker/spiders/). (5) The results (e.g., segmentation volumes and final report in Fig. 3e) are pushed back to XNAT database